Chaperones, Canalization, and Evolution of Animal Forms
Abstract
:1. Robustness and Evolutionary theory
2. Canalization
2.1. Genetic Canalization
2.2. Environmental Canalization
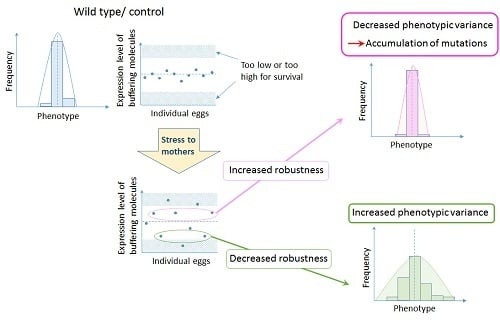
3. Importance of Maternal Control in Canalization of Animal Forms
4. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HSP | Heat shock protein |
| GFP | Green fluorescent protein |
References
- Willmund, F.; Alamo, M.D.; Pechmann, S.; Chen, T.; Albanese, V.; Dammer, E.B.; Peng, J.; Fryman, J. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 2013, 152, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E. Heat shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Morley, J.F.; Morimoto, R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 2004, 15, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Labbadia, J.; Morimoto, R.I. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell 2015, 59, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Casanueva, M.O.; Burga, A.; Lehner, B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science 2012, 335, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wagner, A. Hsp90 is important for fecundity, longevity, and buffering of cryptic deleterious variation in wild fly populations. BMC Evol. Biol. 2012, 12, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, E.J.; Baldwin, L.A. Toxicology rethink its central belief. Nature 2003, 421, 691–692. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, T. A cybernetic view of biological growth. The Maia Hypothesis. Cambridge University Press: Cambridge, UK, 2011; ISBN 9780511933813. [Google Scholar]
- Kishimoto, S.; Uno, M.; Okabe, E.; Nono, M.; Nishida, E. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Comm. 2017, 8, 14031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klosin, A.; Lehner, B. Mechanisms, timescales and principles of trans-generational epigenetic inheritance in animals. Curr. Opin. Genet. Dev. 2016, 36, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Waddington, C.H. The Strategy of the Genes; George Allen & Unwin: London, UK, 1957. [Google Scholar]
- Gibson, G.; Wagner, A. Canalization in evolutionary genetics: A stabilizing theory? BioEssays 2000, 22, 372–380. [Google Scholar] [CrossRef]
- Rutherford, S.L.; Lindquist, S. Hsp90 as a capacitor for morphological variation. Nature 1998, 396, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Lachowiec, J.; Queitsch, C.; Kliebenstein, D.J. Molecular mechanisms governing different robustness of development and environmental responses in plants. Ann. Bot. 2016, 117, 759–809. [Google Scholar] [CrossRef] [PubMed]
- Siegal, M.L.; Leu, J.-Y. On the nature and evolutionary impact of phenotypic robustness mechanisms. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 496–517. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.S.; Kawecki, T.J. Fitness sensitivity and the canalization of life-history traits. Evolution 1994, 48, 1438–1450. [Google Scholar]
- Dworkin, I. Canalization, cryptic variation, and developmental buffering: A critical examination and analytical perspective. In Variation; Hallgrimsson, B., Hall, B., Eds.; Elsevier: Berlington, VT, USA, 2005; pp. 131–158. [Google Scholar]
- Félix, M.-A.; Barkoulas, M. Pervasive robustness in biological systems. Nat. Rev. Genet. 2015, 16, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K. Phenotypic plasticity and robustness: Evolutionary stability theory. Evolutionary Systems Biology. Adv. Exp. Med. Biol. 2012, 751, 249–278. [Google Scholar] [PubMed]
- Bateson, P.; Gluckman, P. Plasticity, Robustness, Development and Evolution; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- West-Eberhard, M.J. Developmental Plasticity and Evolution. Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- DeWitt, T.J.; Scheiner, S.M. Phenotypic Plasticity. Functional and Conceptual Approach; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Wagner, A. Robustness and evolvability: A paradox resolved. Proc. Biol. Sci. 2008, 275, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draghi, J.A.; Parsons, T.L.; Wagner, G.P.; Plotkin, J.B. Mutational robustness can facilitates adaptation. Nature 2010, 463, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Meiklejohn, C.D.; Hartl, D.L. A single mode of canalization. Trends Ecol. Evol. 2002, 17, 468–473. [Google Scholar] [CrossRef]
- Wagner, G.P.; Booth, G.; Bagheri-Chaichian, H. A population genetic theory of canalization. Evolution 1997, 51, 329–347. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K. Evolution of robustness to noise and mutation in gene expression dynamics. PLoS ONE 2007, 5, e434. [Google Scholar] [CrossRef] [PubMed]
- Siegal, M.L.; Bergman, A. Waddington’s canalization revisited: Developmental stability and evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 10528–10532. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A. Genotype networks shed light on evolutionary constraints. Trends Ecol. Evol. 2011, 26, 577–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queitsch, C.; Sangster, T.A.; Lindquist, S. Hsp90 as a capacitor of phenotypic variation. Nature 2002, 6, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, D.F.; Lindquist, S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 2010, 330, 1820–1824. [Google Scholar] [CrossRef] [PubMed]
- Yeyati, P.L.; Bancewicz, R.M.; Maule, J.; van Heyningen, V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007, 30, e43. [Google Scholar]
- Rohner, N.; Jarosz, D.F.; Kowalko, J.E.; Yoshizawa, M.; Jeffery, W.R.; Borowsky, R.L.; Lindquist, S.; Tabin, C.J. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 2013, 342, 1372–1375. [Google Scholar] [CrossRef] [PubMed]
- Geller, R.; Pechmann, S.; Acevedo, A.; Andino, R.; Frydman, J. Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation. Nat. Comm. 2018, 9, 1781. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Tawfick, D. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 2009, 459, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Rodringuez, J.; Sabater-Munoz, B.; Montagud-Matinez, R.; Berlanga, V.; Alvarez-Ponce, D.; Wagner, A.; Fares, M.A. The molecular chaperone DnaK is a source of mutational robustness. Genom. Biol. Evol. 2016, 5, 2979–2991. [Google Scholar] [CrossRef] [PubMed]
- Maisnier-Patin, S.; Roth, J.R.; Fredriksson, A.; Nyström, T.; Berg, O.G.; Andersson, D.I. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat. Genet. 2005, 37, 1376–1379. [Google Scholar] [CrossRef] [PubMed]
- Milton, C.C.; Huynh, B.; Batterham, P.; Rutherford, S.L.; Hoffmann, A.A. Quantitative trait symmetry dependent of Hsp90 buffering: Distinct modes of genetic canalization and developmental stability. Proc. Natl. Acad. Sci. USA. 2003, 100, 13396–13401. [Google Scholar] [CrossRef] [PubMed]
- Milton, C.C.; Batterham, P.; McKenzie, J.A.; Hoffmann, A.A. Effect of E(sev) and Su(Raf) Hsp83 mutants and trans-heterozygous on bristle trait means and variation in Drosophila melanogaster. Genetics 2005, 171, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Debat, V.; Milton, C.C.; Rutherford, S.; Klingenberg, C.P.; Hoffmann, A.A. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution 2006, 60, 2529–2538. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’Alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 2010, 463, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Geiler-Samerotte, K.A.; Zhu, Y.O.; Goulet, B.E.; Hall, D.W.; Siegal, M.L. Selection transforms the landscape of genetic variation interacting with Hsp90. PLoS Biol. 2016, 14, e2000465. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.B.; Uppendahl, L.D.; Traficante, M.K.; Levy, S.F.; Siegal, M.L. Histone variant HTZ1 shows extensive epistasis with, but does not increase robustness to, new mutations. PLoS Genet. 2013, 9, e1003733. [Google Scholar] [CrossRef] [PubMed]
- Schell, R.; Mullis, M.; Ehrenrich, I.M. Modifiers of the genotype-phenotype map: Hsp90 and beyond. PLoS Biol. 2016, 14, e2001015. [Google Scholar] [CrossRef] [PubMed]
- Geiler-Samerotte, K.A.; Sartori, F.M.O.; Siegal, M.L. Decanalizing thinking on genetic canalization. Semin. Cell Dev. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Paaby, A.; White, A.G.; Riccardi, D.D.; Gunsalus, K.C.; Piano, F.; Rockman, M.V. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife 2015, 22, 4. [Google Scholar] [CrossRef] [PubMed]
- Milloz, J.; Duveau, F.; Nuez, I.; Felix, M.-A. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008, 22, 3064–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsanos, D.; Koneru, S.L.; Boukhibar, L.M.; Gritti, N.; Chose, R.; Appleford, P.J.; Doitsidou, M.; Woollard, A.; van Zon, J.S.; Poole, R.J.; et al. Stochastic loss and gain of symmetric divisions in the C. elgans epidermis perturbs robustness of stem cell number. PLoS Biol. 2017, 2017, 2002429. [Google Scholar] [CrossRef]
- Cassidy, J.J.; Jha, A.R.; Posadas, D.M.; Giri, R.; Venken, K.J.; Ji, J.; Jiang, H.; Bellen, H.J.; White, K.I.; Carthew, R.J. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell 2013, 155, 1556–1567. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.J.; Straughan, A.J.; Carthew, R.W. Differential masking of natural genetic variation by miR-9a in Drosophila. Genetics 2016, 202, 675687. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.H. Multiple capacitors for natural genetic variation in Drosophila melanogaster. Mol. Ecol. 2013, 22, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.H. Novel genetic capacitors and potentiators for the natural genetic variation of sensory bristles and their trait specificity in Drosophila melanogaster. Mol. Ecol. 2015, 24, 5561–5572. [Google Scholar] [CrossRef] [PubMed]
- Lehner, B.; Crombie, C.; Tischler, J.; Fortunato, A.; Fraser, A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006, 38, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Rinott, R.; Jamovich, A.; Friedman, N. Exploring transcription regulation through cell-to-cell variability. Proc. Natl. Acad. Sci. USA 2011, 108, 6329–6334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, A.A.; Willi, Y. Detecting genetic responses to environmental change. Nat. Rev. Genet. 2008, 9, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Kawashima, T.; Fujie, M.; Hughes, S.; Satoh, N.; Shimeld, S. Molecular basis of canalization in an ascidian species complex adapted to different thermal conditions. Sci. Rep. 2015, 5, 16717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.M.; Nishikawa, T.; Bird, A. Genomic approaches reveal unexpected genetic divergence within Ciona intestinalis. J. Mol. Evol. 2005, 61, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Andreakis, N.; Mastrototaro, F.; Cirino, P.; Vassillo, M.; Sordino, P. Cryptic speciation in a model invertebrate chordate. Proc. Natl. Acad. Sci. USA 2007, 104, 9364–9369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iannelli, F.; Pesole, G.; Sordino, P.; Gissi, C. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 2007, 23, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Nydam, M.L.; Harrison, R.G. Genealogical relationships within and among shallow-water Ciona species (Ascidiacea). Mar. Biol. 2007, 151, 1839–1847. [Google Scholar] [CrossRef]
- Zhan, A.; Macisaac, H.J.; Cristescu, M.E. Invasion genetics of the Ciona intestinalis species complex: From regional endemism to global homogeneity. Mol. Ecol. 2010, 19, 4678–4694. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Satoh, N.; Bishop, J.D.D. Field identification of ‘types’ A and B of the ascidian Ciona intestinalis in a region of sympatry. Mar. Biol. 2012, 159, 1611–1619. [Google Scholar] [CrossRef]
- Sato, A.; Shimeld, S.M.; Bishop, J.D.D. Symmetrical reproductive compatibility of two species in the Ciona intestinalis (Ascidiacea) species complex, a model for marine genomics and developmental biology. Zool. Sci. 2014, 31, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Boukhibar, L.M.; Barkoulas, M. The developmental genetics of biological robustness. Ann. Bot. 2016, 117, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Ushioda, R.; Hoseki, J.; Araki, K.; Jansen, G.; Thomas, D.Y.; Nagata, K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008, 321, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Futschik, M.E.; Teichmann, S.A.; Babu, M.M. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science 2008, 322, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Loison, L. Canalization and genetic assimilation: Reassessing the radicality of the Waddingtonian concept of inheritance of acquired characters. Semin. Cell Dev. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Marszalek, J. How do J-proteins get Hsp70 to do so many different things? Trend Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, H. Temperature sensitivity of a hybrid between two species of sea urchin differing in thermotolerance. Dev. Growth Differ. 1993, 35, 395–401. [Google Scholar] [CrossRef]
- Kubota, Y.; Shima, A. Comparative study of embryonic thermoresistance of two inbred strains of the medaka (Oryzias latipes). J. Comp. Physiol. B 1991, 160, 621–625. [Google Scholar] [CrossRef]
- Kusnadi, E.P.; Hannan, K.M.; Hicks, R.J.; Hannan, R.D.; Pearson, R.B.; Kang, J. Regulation of rDNA transcription in response to growth factors, nutrients and energy. Gene 2015, 556, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Denienko, O.; Lucas, E.S.; Congshan, S.; Watkins, A.J.; Mar, D.; Bomsztyk, K.; Flemming, T.P. Regulation of ribosomal RNA expression across the lifespan is fine-tuned by maternal diet before implantation. Biochem. Biophys. Acta 2016, 1859, 906–913. [Google Scholar] [CrossRef] [Green Version]
- Blake, G.E.T.; Watson, E.D. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr. Opin. Chem. Biol. 2016, 33, 101–107. [Google Scholar] [CrossRef] [PubMed]
- O’Dea, R.E.; Noble, D.W.A.; Johnson, S.L.; Hesselson, D.; Nakagawa, S. The role of non-genetic inheritance in evolutionary rescue: Epigenetic buffering, heritable bet-hedging and epigenetic traps. Environ. Epigenet. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.J.; Spencer, H.G.; Donohue, K.; Sultan, S.E. How stable ‘should’ epigenetic modification be? Insights from adaptive plasticity and bet hedging. Evolution 2013, 68, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Nishida, H. REGULATOR: A database of metazoan transcription factors and maternal factors for developmental studies. BMC Bioinform. 2016, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Dennis, L. Patterns of synthesis and accumulation of heterogeneous RNA in lampbrush stage oocytes of Xenopus laevis (Daudin). Dev. Biol. 1978, 67, 274–285. [Google Scholar] [CrossRef]
- Golden, L.; Schafer, U.; Rosbash, M. Accumulation of individual pA+ RNAs during oogenesis of Xenopus laevis. Cell 1980, 22, 835–844. [Google Scholar] [CrossRef]
- Capco, D.; Jeffery, W.R. Origin and spatial distribution of maternal messenger RNA during oogenesis of an insect, Oncopeltus fasciatus. J. Cell Sci. 1979, 39, 63–76. [Google Scholar] [PubMed]
- Davenport, R. Transport of ribosomal RNA into the oöcytes of the milkweed bug, Oncopeltus fasciatus. J. Insect Physiol. 1976, 22, 925–926. [Google Scholar] [CrossRef]
- Saxton, W.M. Microtubles, motors, and mRNA localization mechanisms: Watching fluorescent message move. Cell 2001, 107, 707–710. [Google Scholar] [CrossRef]
- St Johnston, D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005, 6, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Walser, C.B.; Lipshitz, H.D. Transcript clearance during the maternal-to-zygotic transition. Curr. Opin. Genet. Dev. 2011, 21, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, S.; Andres, S.; Janga, S.C.; Huber, W.; Alonso, C.R. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genom. Biol. 2010, 11, R93. [Google Scholar] [CrossRef] [PubMed]
- Geisberg, J.V.; Moqtaderi, Z.; Fan, X.; Ozsolak, F.; Struhl, K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 2014, 156, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Peshkin, L.; Wuhr, M.; Pearl, E.; Haas, W.; Freeman, R.M.; Gerhart, J.C.; Klein, A.M.; Horb, M.; Gygi, S.P.; Kirschner, W.M. On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Dev. Cell 2015, 35, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Hynes, R.; Kirschner, M. Temporal and spatial regulation of fibronectin in early Xenopus development. Cell 1984, 36, 729–740. [Google Scholar] [CrossRef]
- Crean, A.J.; Marshall, D.J. Coping with environmental uncertainty: Dynamic bet hedging as a maternal effect. Philos. Trans. Biol. Sci. 2009, 364, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Simons, A.M. Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc. Biol. Sci. 2011, 278, 1601–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaumont, H.J.E.; Gallie, J.; Kost, C.; Ferguson, G.C.; Rainey, R.B. Experimental evolution of bet hedging. Nature 2009, 462, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.F.; Ziv, N.; Siegal, M.L. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012, 10, E1001325. [Google Scholar] [CrossRef]
- Danforth, B.N. Emergence dynamics and bet hedging in a desert bee, Perdita partalis. Proc. Biol. Sci. 1999, 266, 1985–1994. [Google Scholar] [CrossRef]
- López-Maury, L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.F.; Siegal, M.L. Evolutionary systems biology, advances in experimental medicine and biology. In The Robustness Continuum; Soyer, O.S., Ed.; Springer New York: New York, NY, USA; Volume 751, pp. 431–452.
- Lockwood, B.L.; Julick, C.R.; Montooth, K.L. Maternal loading of a small heat shock proteinin creases embryo thermal tolerance in Drosophila melanogaster. J. Exp. Biol. 2017, 220, 4492–4501. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.; Snir, O.; Mizrachi, E.; Galili, M.; Zaltsman, I.; Soen, Y. Reduction in maternal Polycomb levels contributes to transgenerational inheritance of a response to toxic stress in flies. J. Physiol. 2014, 592, 2243–2355. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, A. Chaperones, Canalization, and Evolution of Animal Forms. Int. J. Mol. Sci. 2018, 19, 3029. https://doi.org/10.3390/ijms19103029
Sato A. Chaperones, Canalization, and Evolution of Animal Forms. International Journal of Molecular Sciences. 2018; 19(10):3029. https://doi.org/10.3390/ijms19103029
Chicago/Turabian StyleSato, Atsuko. 2018. "Chaperones, Canalization, and Evolution of Animal Forms" International Journal of Molecular Sciences 19, no. 10: 3029. https://doi.org/10.3390/ijms19103029
APA StyleSato, A. (2018). Chaperones, Canalization, and Evolution of Animal Forms. International Journal of Molecular Sciences, 19(10), 3029. https://doi.org/10.3390/ijms19103029