Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures
Abstract
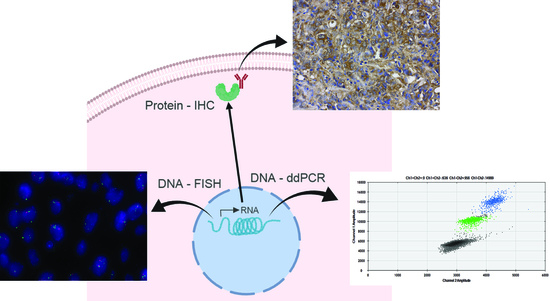
:1. Introduction
2. Results
2.1. Immunohistochemistry Analysis Demonstrates Variable Marker Expression between MPM Tissue and Derivative Cell-Line Samples
2.2. Genomic Deletion of CDKN2A Was Identified in MPM Samples Using FISH
2.3. Copy Number Variation Contributes to Loss of BAP1 and CDKN2A Expression in MPM
2.4. Concordance of BAP1 Protein Expression and Genomic Deletion
3. Discussion
4. Materials and Methods
4.1. Patient Tissue-Sample Collection and MPM Cell-Line Establishment
MPM Cell-Line Establishment
4.2. Immunohistochemical Analysis of MPM Tissue Sections and Established Cell-Line Blocks
4.3. FISH Analysis of CDKN2A Genomic Analysis
4.4. CDKN2A and BAP1 Genomic Loss Were Suggested by CNV Using ddPCR
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Robinson, B.W.; Lake, R.A. Advances in malignant mesothelioma. N. Engl. J. Med. 2005, 353, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- LaDou, J.; Castleman, B.; Frank, A.; Gochfeld, M.; Greenberg, M.; Huff, J.; Joshi, T.K.; Landrigan, P.J.; Lemen, R.; Myers, J.; et al. The case for a global ban on asbestos. Environ. Health Perspect. 2010, 118, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Carbone, M.; Kanodia, S.; Chao, A.; Miller, A.; Wali, A.; Weissman, D.; Adjei, A.; Baumann, F.; Boffetta, P.; Buck, B.; et al. Consensus Report of the 2015 Weinman International Conference on Mesothelioma. J. Thorac. Oncol. 2016, 11, 1246–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kao, S.C.; Reid, G.; Lee, K.; Vardy, J.; Clarke, S.; van Zandwijk, N. Malignant mesothelioma. Intern. Med. J. 2010, 40, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Tsao, A.S.; Wistuba, I.; Roth, J.A.; Kindler, H.L. Malignant pleural mesothelioma. J. Clin. Oncol. 2009, 27, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.H.; Schiller, J.H.; Bunn, P.A., Jr. Recent clinical advances in lung cancer management. J. Clin. Oncol. 2014, 32, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Scherpereel, A.; Astoul, P.; Baas, P.; Berghmans, T.; Clayson, H.; de Vuyst, P.; Dienemann, H.; Galateau-Salle, F.; Hennequin, C.; Hillerdal, G.; et al. Guidelines of the European Respiratory Society and the European Society of Thoracic Surgeons for the management of malignant pleural mesothelioma. Eur. Respir. J. 2010, 35, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Linton, A.; Kao, S.; Vardy, J.; Clarke, S.; van Zandwijk, N.; Klebe, S. Immunohistochemistry in the diagnosis of malignant pleural mesothelioma: Trends in Australia and a literature review. Asia Pac. J. Clin. Oncol. 2013, 9, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.N.; Colby, T.; Ordonez, N.; Krausz, T.; Attanoos, R.; Beasley, M.B.; Borczuk, A.C.; Butnor, K.; Cagle, P.T.; Chirieac, L.R.; et al. Guidelines for pathologic diagnosis of malignant mesothelioma: 2012 Update of the consensus statement from the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2013, 137, 647–667. [Google Scholar] [CrossRef] [PubMed]
- Tischoff, I.; Neid, M.; Neumann, V.; Tannapfel, A. Pathohistological diagnosis and differential diagnosis. Recent Results Cancer Res. 2011, 189, 57–78. [Google Scholar] [PubMed]
- Oehl, K.; Vrugt, B.; Opitz, I.; Meerang, M. Heterogeneity in Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2018, 19, 1603. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Chmielecki, J.; Goparaju, C.; Heguy, A.; Dolgalev, I.; Carbone, M.; Seepo, S.; Meyerson, M.; Pass, H.I. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015, 75, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Hylebos, M.; Van Camp, G.; van Meerbeeck, J.P.; Op de Beeck, K. The Genetic Landscape of Malignant Pleural Mesothelioma: Results from Massively Parallel Sequencing. J. Thorac. Oncol. 2016, 11, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Kim, D.; Hong, Y.S.; Kim, K.P.; Yoon, Y.K.; Lee, D.H.; Kim, S.W.; Chun, S.M.; Jang, S.J.; Kim, T.W. Mutational Profiling of Malignant Mesothelioma Revealed Potential Therapeutic Targets in EGFR and NRAS. Transl. Oncol. 2018, 11, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Jaurand, M.C.; Fleury-Feith, J. Pathogenesis of malignant pleural mesothelioma. Respirology 2005, 10, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borczuk, A.C.; Pei, J.; Taub, R.N.; Levy, B.; Nahum, O.; Chen, J.; Chen, K.; Testa, J.R. Genome-wide analysis of abdominal and pleural malignant mesothelioma with DNA arrays reveals both common and distinct regions of copy number alteration. Cancer Biol. Ther. 2016, 17, 328–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, A.V.; Goparaju, C.M.; Ivanov, S.V.; Nonaka, D.; Cruz, C.; Beck, A.; Lonardo, F.; Wali, A.; Pass, H.I. Protumorigenic role of HAPLN1 and its IgV domain in malignant pleural mesothelioma. Clin. Cancer Res. 2009, 15, 2602–2611. [Google Scholar] [CrossRef] [PubMed]
- Illei, P.B.; Rusch, V.W.; Zakowski, M.F.; Ladanyi, M. Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin. Cancer Res. 2003, 9, 2108–2113. [Google Scholar] [PubMed]
- Dahabreh, I.J.; Linardou, H.; Siannis, F.; Kosmidis, P.; Bafaloukos, D.; Murray, S. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2010, 16, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Nasu, M.; Emi, M.; Pastorino, S.; Tanji, M.; Powers, A.; Luk, H.; Baumann, F.; Zhang, Y.A.; Gazdar, A.; Kanodia, S.; et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J. Thorac. Oncol. 2015, 10, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Cigognetti, M.; Lonardi, S.; Fisogni, S.; Balzarini, P.; Pellegrini, V.; Tironi, A.; Bercich, L.; Bugatti, M.; Rossi, G.; Murer, B.; et al. BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod. Pathol. 2015, 28, 1043–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheffield, B.S.; Hwang, H.C.; Lee, A.F.; Thompson, K.; Rodriguez, S.; Tse, C.H.; Gown, A.M.; Churg, A. BAP1 immunohistochemistry and p16 FISH to separate benign from malignant mesothelial proliferations. Am. J. Surg. Pathol. 2015, 39, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Sato, G. Tissue culture: The unrealized potential. Cytotechnology 2008, 57, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Sato, G.H.; Sato, J.D.; Okamoto, T.; McKeehan, W.L.; Barnes, D.W. Tissue culture: The unlimited potential. In Vitro Cell. Dev. Biol. Anim. 2010, 46, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.V.; Ivanova, A.V.; Goparaju, C.M.; Chen, Y.; Beck, A.; Pass, H.I. Tumorigenic properties of alternative osteopontin isoforms in mesothelioma. Biochem. Biophys. Res. Commun. 2009, 382, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.S.K.; Lee, K.; Clarke, C.J.; Cheng, N.C.; van Zandwijk, N.; Klebe, S.; Reid, G. Establishing malignant pleural mesothelioma cell lines using the spheroid method produces a model with better 3D architecture. J. Thorac. Oncol. 2017, 12, S2265–S2266. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Sato, A.; Tsujimura, T.; Emi, M.; Morinaga, T.; Fukuoka, K.; Yamada, S.; Murakami, A.; Kondo, N.; Matsumoto, S.; et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 2012, 103, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.; Testa, J.R. BAP1, a tumor suppressor gene driving malignant mesothelioma. Transl. Lung Cancer Res. 2017, 6, 270–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Port, J.; Murphy, D.J. Mesothelioma: Identical Routes to Malignancy from Asbestos and Carbon Nanotubes. Curr. Biol. 2017, 27, R1173–R1176. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.E.; Karrison, T.; Ananthanarayanan, V.; Gallan, A.J.; Adusumilli, P.S.; Alchami, F.S.; Attanoos, R.; Brcic, L.; Butnor, K.J.; Galateau-Sallé, F.; et al. Nuclear grade and necrosis predict prognosis in malignant epithelioid pleural mesothelioma: A multi-institutional study. Mod. Pathol. 2018, 31, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Almendro, V.; Polyak, K. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer 2012, 12, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.C.; Klebe, S.; Henderson, D.W.; Reid, G.; Chatfield, M.; Armstrong, N.J.; Yan, T.D.; Vardy, J.; Clarke, S.; van Zandwijk, N.; et al. Low calretinin expression and high neutrophil-to-lymphocyte ratio are poor prognostic factors in patients with malignant mesothelioma undergoing extrapleural pneumonectomy. J. Thorac. Oncol. 2011, 6, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Marchevsky, A.M. Application of immunohistochemistry to the diagnosis of malignant mesothelioma. Arch. Pathol. Lab. Med. 2008, 132, 397–401. [Google Scholar] [PubMed]
- Relan, V.; Morrison, L.; Parsonson, K.; Clarke, B.E.; Duhig, E.E.; Windsor, M.N.; Matar, K.S.; Naidoo, R.; Passmore, L.; McCaul, E.; et al. Phenotypes and karyotypes of human malignant mesothelioma cell lines. PLoS ONE 2013, 8, e58132. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Mohan, D.K.; Sahu, B. Protein Expression Differences of 2-Dimensional and Progressive 3-Dimensional Cell Cultures of Non-Small-Cell-Lung-Cancer Cell Line H460. J. Cell. Biochem. 2017, 118, 1648–1652. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, K.; Matsumoto, S.; Hamasaki, M.; Hida, T.; Kamei, T.; Hiroshima, K.; Tsujimura, T.; Kawahara, K. Use of p16 FISH for differential diagnosis of mesothelioma in smear preparations. Diagn. Cytopathol. 2016, 44, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Riegel, M. Human molecular cytogenetics: From cells to nucleotides. Genet. Mol. Biol. 2014, 37, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Mok, E.; Tan, S.; Leygo, C.; McLaughlin, C.; George, A.M.; Reid, G. SFRP Tumour Suppressor Genes Are Potential Plasma-Based Epigenetic Biomarkers for Malignant Pleural Mesothelioma. Dis. Mark. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hida, T.; Hamasaki, M.; Matsumoto, S.; Sato, A.; Tsujimura, T.; Kawahara, K.; Iwasaki, A.; Okamoto, T.; Oda, Y.; Honda, H.; et al. BAP1 immunohistochemistry and p16 FISH results in combination provide higher confidence in malignant pleural mesothelioma diagnosis: ROC analysis of the two tests. Pathol. Int. 2016, 66, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.T.; Santos Gda, C.; Hwang, D.M.; Ludkovski, O.; Pintilie, M.; Squire, J.A.; Tsao, M.-S. FISH assay development for the detection of p16/CDKN2A deletion in malignant pleural mesothelioma. J. Clin. Pathol. 2010, 63, 630–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| MM ID | Sample Type | CK 8/18 | Calretinin | CK 5/6 | CD141 | HBME-1 | WT-1 | D2-40 | EMA | CEA | TAG72 | BG8 | CD15 | TTF-1 | BAP1 | Ber-EP4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1137 | Tissue | (+++) 90% | - | (+++) <5% | (++) 10% | - | (++) 10% | (+) <5% | - | - | - | (++) 70% | - | - | (+++) 90–100% | - |
| 1137 T | 3D cells | (+++) 100% | - | - | - | - | - | - | - | - | - | (++/+++) 90–100% | - | - | (+++) 30% | - |
| 2D cells | (+++) 90% | (++) 5% | (+) 10% | (+) 20% | (+++) 10% | (+) 10% | - | (+) <5% | (++) 10% | - | (++) 60% | - | (+++) <5% | - | (++/+++) 60% | |
| 1157 | tissue | (+++) 90–100% | (+++) 80% | (+++) 90–100% | (+) 50% | (+++) 10% | (++) 90% | (++) 30% | (+++) 40% | - | - | (+++) 90–100% | - | - | - | - |
| 1157 T | 3D cells | (+++) 100% | (+) 40% | (+++) 70% | - | - | (+++) 90% | (+) <10% | (+++) 40% | - | - | (+) 40% | - | - | - | - |
| 2D cells | (+++) 90–100% | - | (++/+++) 90% | (+) <5% | - | - | - | (+) <5% | - | - | - | - | - | - | (+) 50% | |
| 1180 | Tissue | (++++) 90–100% | (+++) 30% | (+++) <5% | (+++) 30% | (+++) <5% | - | (+++) 10% | - | - | - | (++) 80% | - | - | (+++) 80% | - |
| 1180 T | 3D cells | (+++) 100% | - | - | - | - | (++) 30% | - | (++) 40% | - | - | (+) 30% | - | (+) 10% | (+++) 95% | - |
| 2D cells | (+++) 90–100% | - | - | - | - | (+++) <10% | - | - | - | - | - | - | (++) <10% | (+++) 95% | - | |
| 1187 | tissue | (+++) 90–100% | (+++) 90–100% | (+++) 90–100% | (+++) 90–100% | (+++) 90–100% | (+++) 90–100% | (+++) 40% | - | - | (+++) 90–100% | - | - | (++) 90–100% | - | (++) 90–100% |
| 1187 T | 3D cells | (++) 10% | - | - | - | - | (+++) 90% | (++) 40% | - | - | - | - | - | - | (+++) 80% | - |
| 2D cells | (+++) 100% | - | - | (++) 40% | - | (++) 80% | (+) 10% | - | - | - | (+) 70% | - | - | (+++) 95% | (+) 80% | |
| 1505 | Tissue | (+++) 90–100% | (+++) 40% | (+++) 10% | (++) 30% | - | (++) <5% | (+++) <5% | - | - | - | (+++) 90–100% | - | (+++) 40% | (+++) 90–100% | - |
| 1505 T | 3D cells | (+++) 100% | (+) 20% | (+) <5% | (+) <5% | (+) <5% | (+) <5% | - | (++) 60% | (++) 60% | (+) 20% | (+++) 100% | - | (+++) 80% | (+++) 100% | (+++) 50% |
| 2D cells | (+++) 100% | (++) 80% | (+) 80% | (+) 70% | (+) 20% | - | (+) 30% | - | (+) <5% | (++) 60% | (+++) 100% | (+) 80% | (+++) 80% | (+++) 100% | (+++) 100% | |
| 1506 | Tissue | (+++) 90% | (+++) 80% | - | (++) 70% | (+++) 100% | (++) 60% | (+) 10% | - | - | - | (++) 40% | - | - | (+++) 90% | (+) 10% |
| 1506 T | 3D cells | - | - | (+) <5% | (+++) 40% | - | - | - | (++) 80% | - | - | (+++) 90% | - | - | (+++) 100% | (+++) 80% |
| 2D cells | (+) 10% | - | (+) 40% | (+) 10% | (+) <5% | - | - | (++) 40% | (+) 20% | (++) 70% | (+++) 100% | (+) 90% | (+) <5% | (+++) 80% | (+++) 100% | |
| 1518 | Tissue | (+++) 40% | - | (+++) 30% | (+) 20% | - | (++) 10% | (++) 30% | (++) 10% | - | - | (+) 40% | - | - | (+++) 20% | - |
| 1518 P | 3D cells | (+++) 100% | (+) 10% | (+) <5% | - | (+) 5% | (+++) 40% | (+) <5% | (+) <5% | - | - | (+++) 100% | - | - | (+++) 90% | (+++) 90% |
| 2D cells | (+++) 100% | (+) 5% | - | (+) 10% | - | (+) 10% | (+) <5% | (+) <5% | (+) 10% | - | (+++) 100% | (+) 80% | (++) 80% | (+++) 100% | ||
| 1518 T | 3D cells | (+++) 70% | (++) 10% | - | (+) <5% | - | - | - | - | - | - | - | - | - | - | (+) 10% |
| 2D cells | - | (++) 4% | - | (+) <5% | - | (+++) 40% | (++) 30% | - | (++/+++) 10% | (+) 10% | - | - | (+++) 20% | - | (+++) 40% | |
| 1170 T | 3D cells | (++/+++) 70% | - | - | (+) 40% | (++) <5% | (+++) 20% | - | (+++) 30% | - | - | (+) 10% | (+) <5% | (+++) 90–100% | (+++) 95% | (++/+++) 40% |
| 2D cells | (++) 70% | - | - | - | (++) 70% | (++/+++) 80% | - | - | - | - | - | - | (+++) 80% | (+++) 95% | (++) 20% | |
| 1843 | Tissue | (++) 40% | (+) <10% | (+++) 40% | (+++) 30% | (+++) 30% | (+) 5% | - | (+++) 20% | - | - | (+) 20% | - | (+) <5% | - | - |
| 1843 T | 3D cells | (+++) 50% | - | - | - | - | (+++) 100% | (+) 20% | (++/+++) 40% | - | - | - | - | (+++) 90% | - | - |
| 2D cells | (++) 80% | - | - | - | - | (++) 70% | - | - | - | - | (+) 50% | - | (++) 80% | - | (+) 50% | |
| 2164 | Tissue | (+++) 40% | (++) 40% | - | (++) 40% | - | (+) <5% | (+++) 70% | (+++) 60% | - | - | (+) 10% | - | - | - | - |
| 2164 P | 3D cells | - | - | - | - | - | (++) 90% | - | - | - | - | - | - | - | - | - |
| 2D cells | - | - | - | - | - | (++) 90–100% | (++) 30% | (+) 10% | - | - | (+) 40% | - | - | - | - | |
| 2170 | Tissue | (+++) 90% | (+++) 60% | (+) 5% | (++) 40% | (+++) 90% | (+++) 40% | (+++) 80% | (+++) 80% | - | - | (++) 10% | - | - | - | (+++) 5% |
| 2170 T | 3D cells | - | (+) <5% | - | - | - | - | - | - | (+) 15% | - | (+++) 100% | (++) 5% | - | (+++) 100% | - |
| 2D cells | - | - | - | (++) 10% | (+) 5% | - | (+) <5% | (+) 10% | - | - | (+++) 100% | (+) 10% | - | (+++) 100% | (+) 80% | |
| 2174 P | 3D | (+++) 100% | (++) <5% | - | (+++) 50% | (+++) 100% | (+++) 80% | - | (++) 40% | - | - | (+++) 100% | - | - | (+++) 100% | - |
| 2D | (+++) 80% | (++) 80% | - | (+++) 90% | (+) 20% | - | (+++) 90% | (+++) 80% | (+) 10% | - | (+++) 100% | (+) 90% | - | (+++) 70% | (+) 90% | |
| 2175 | Tissue | (+++) 100% | (+++) 60% | (+++80%) | - | (+) 10% | (+) 20% | (+) 50% | (+++) 70% | - | - | - | - | - | (+++) 80% | - |
| 2175 P | 3D cells | - | - | - | (+++) 60% | - | - | - | (++/+++) 80% | - | - | - | - | - | (+++) 95% | - |
| 2D cells | - | - | - | (++) <5% | - | - | - | (+) <5% | - | - | - | - | - | (+++) 95% | - | |
| 2280 | Tissue | (+++) 100% | (+) <5% | - | (+++) 90% | - | - | - | (+) <5% | - | - | - | - | - | (+) 100% | - |
| 2280 T | 3D cells | - | - | - | (+++) 60% | - | - | - | (+++) 80% | - | - | (+++) 80% | - | - | (+++) 100% | - |
| 2D cells | (+++) <5% | - | - | (+) 5% | - | - | (+) <5% | (+) 10% | - | - | (+++) 100% | (+) 20% | - | (+++) 100% | (+) 90% |
| MM ID | Sample Type | BAP1 IHC | BAP1 ddPCR (Reference to RPP30) |
|---|---|---|---|
| 1137 | Tissue | (+++) 90–100% | 1.94 |
| cells | (+++) 30% | 2 | |
| 1157 | Tissue | ND | 0.028 |
| cells | ND | 0.07 | |
| 1180 | Tissue | (+++) 80% | 2.91 |
| cells | (+++) 95% | 1.69 | |
| 1187 | Tissue | ND | 3.28 |
| cells | (+++) 80% | 2 | |
| 1505 | Tissue | (+++) 90–100% | 1.87 |
| cells | (+++) 100% | 2.01 | |
| 1506 | Tissue | (+++) 90% | 1.96 |
| cells | (+++) 100% | 1.87 | |
| 1518 | Tissue | (+++) 20% | 0.82 |
| cells | (+++) 90% | 0.86 | |
| 1170 | Tissue | (+++) 95% | No tissue availible |
| Cells | (+++) 95% | 1.98 | |
| 1843 | Tissue | ND | 1 |
| Cells | ND | 0.94 | |
| 2164 | Tissue | ND | 0.24 |
| Cells | ND | 0.39 | |
| 2170 | Tissue | ND | 1.15 |
| Cells | (+++) 100% | 1 | |
| 2174 | Tissue | No tissue availible | No tissue availible |
| Cells | (+++) 70% | 1.327814 | |
| 2175 | Tissue | (+++) 80% | 1.91 |
| Cells | (+++) 95% | 1.92 | |
| 2280 | Tissue | (+) 100% | 1.88 |
| Cells | (+++) 100% | 1.85 |
| MM ID | Aga at Diagnose | Gender | Histological Subtype | Surgery Procedure | Survival (months) | Asbestos Exposure |
|---|---|---|---|---|---|---|
| 1137 | 83 | Male | Desmoplastic | Biopsy, Decortication, Pleurodesis | 0.7 | Yes |
| 1157 | 51 | Male | Epitheliod | Biopsy, Decortication, Pleurodesis | 20.6 | Yes |
| 1170 | 74 | Male | Biphasic | Biopsy, Decortication, Pleurodesis | 0.8 | Yes |
| 1180 | 72 | Male | Biphasic | Biopsy, Decortication, Pleurodesis | 7.4 | Yes |
| 1187 | 64 | Male | Biphasic | Extrapleural pneumonectomy | 33.5 | Yes |
| 1505 | 57 | Male | Epitheliod | Biopsy, Surgical exploration | 7.2 | Yes |
| 1506 | 72 | Male | Epitheliod | Biopsy, Pleurodesis | 24.0 | Yes |
| 1518 | 80 | Male | Biphasic | Biopsy, Decortication, Pleurodesis | 22.7 | Yes |
| 1843 | 60 | Male | Biphasic | Decortication, Pleurodesis | 15.6 | Yes |
| 2164 | 73 | Male | Epitheliod | Biopsy, Decortication, Pleurodesis | * | Yes |
| 2170 | 75 | Male | Epitheliod | Biopsy, Decortication, Pleurodesis | * | ND |
| 2174 | 78 | Male | Epitheliod | Biopsy, Decortication, Pleurodesis | 3.0 | Yes |
| 2175 | 66 | Male | Epitheliod | Biopsy, Decortication, Pleurodesis | 12.5 | Yes |
| 2280 | 69 | Male | Epitheliod | Extrapleural pneumonectomy | * | Yes |
| Antibody | Clone | Manufacturer | Product Code | Species | Dilution |
|---|---|---|---|---|---|
| CK8/18 | EP17/30 | Dako | M3652 | Rabbit | 1:100 |
| Calretinin | Polyclonal | Biocare | CP092C | Rabbit | 1:100 |
| CK5/6 | D5 & 16B4 | Cell Marque | 358M-16 | Mouse | 1:150 |
| CD141 | 15CB | Novocastra | NCL-CD141 | Mouse | 1:50 |
| HBME1 | HBME-1 | Dako | M3505 | Mouse | 1:50 |
| WT1 | WT49 | Novocastra | NCL-L-WT1-562 | Mouse | 1:50 |
| D2-40 | D2-40 | Biocare | CM266C | Mouse | 1:100 |
| EMA | GP1.4 | Novocastra | NCL-L-EMA | Mouse | 1:350 |
| CEA | 11-7 | Dako | M7072 | Mouse | 1:200 |
| TAG72 | B72-3 | Cell Marque | 337M-84 | Mouse | 1:2000 |
| BG8 | F3 | Covance | SIG-3317-1000 | Mouse | 1:100 |
| CD15 | Carb-3 | Dako | M3631 | Mouse | 1:100 |
| TTF-1 | SPT24 | Novocastra | NCL-L-TTF-1 | Mouse | 1:100 |
| BAP1 | C-4 | Santa Cruz | SC-28383 | Mouse | 1:200 |
| HEA | Ber-EP4 | Dako | M0804 | Mouse | 1:100 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarun, K.H.; Lee, K.; Williams, M.; Wright, C.M.; Clarke, C.J.; Cheng, N.C.; Takahashi, K.; Cheng, Y.Y. Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures. Int. J. Mol. Sci. 2018, 19, 3056. https://doi.org/10.3390/ijms19103056
Sarun KH, Lee K, Williams M, Wright CM, Clarke CJ, Cheng NC, Takahashi K, Cheng YY. Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures. International Journal of Molecular Sciences. 2018; 19(10):3056. https://doi.org/10.3390/ijms19103056
Chicago/Turabian StyleSarun, Kadir Harun, Kenneth Lee, Marissa Williams, Casey Maree Wright, Candice Julie Clarke, Ngan Ching Cheng, Ken Takahashi, and Yuen Yee Cheng. 2018. "Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures" International Journal of Molecular Sciences 19, no. 10: 3056. https://doi.org/10.3390/ijms19103056
APA StyleSarun, K. H., Lee, K., Williams, M., Wright, C. M., Clarke, C. J., Cheng, N. C., Takahashi, K., & Cheng, Y. Y. (2018). Genomic Deletion of BAP1 and CDKN2A Are Useful Markers for Quality Control of Malignant Pleural Mesothelioma (MPM) Primary Cultures. International Journal of Molecular Sciences, 19(10), 3056. https://doi.org/10.3390/ijms19103056