An Ultradian Feeding Schedule in Rats Affects Metabolic Gene Expression in Liver, Brown Adipose Tissue and Skeletal Muscle with Only Mild Effects on Circadian Clocks
Abstract
:1. Introduction
2. Results
2.1. Caloric Intake and Body Weight Gain
2.2. RER
2.3. Locomotor Activity
2.4. Energy Expenditure
2.5. Body Temperature
2.6. Clock and Clock-Controlled Gene Expression in the SCN
2.7. Clock Gene Expression in Soleus Muscle
2.8. Clock Gene Expression in BAT
2.9. Clock Gene Expression in Liver
2.10. Metabolic Genes in Soleus Muscle
2.11. Metabolic Genes in BAT
2.12. Metabolic Genes in Liver
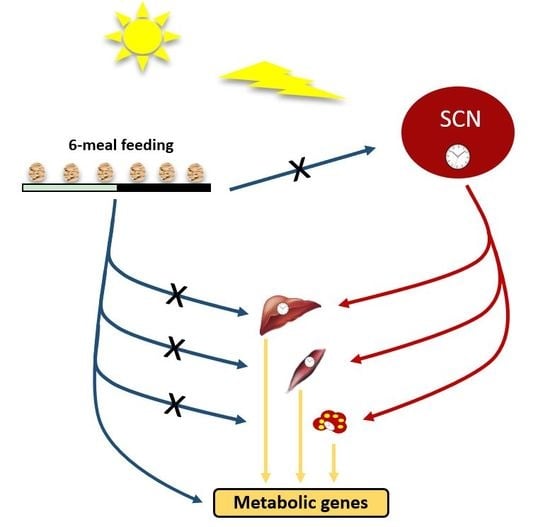
3. Discussion
3.1. Ultradian Rhythms in Feeding Behavior Affect Whole Body Metabolism
3.2. Ultradian Rhythms in Feeding Behavior Do Not Affect the Central SCN Clock
3.3. Ultradian Feeding Behavior Does Not Dictate the Peripheral Clocks
3.4. Ultradian Feeding Behavior Differentially Affects Metabolic Gene Expression
3.4.1. Soleus Muscle
3.4.2. BAT
3.4.3. Liver
3.5. Clinical Relevance of Ultradian and Other TRF Interventions
4. Materials and Methods
4.1. Animal Experiments
4.2. Activity and Respirometry
4.3. Body Temperature
4.4. RNA Isolation
4.5. Muscle, BAT and Liver cDNA Synthesis
4.6. RT-qPCR
4.7. In Situ Hybridization
4.8. Immunohistochemistry
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hastings, M.H.; Reddy, A.B.; Maywood, E.S. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003, 4, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.S.; Young, M.E. The role of cell-specific circadian clocks in metabolism and disease. Obes. Rev. 2009, 10 (Suppl. 2), 6–13. [Google Scholar] [CrossRef] [PubMed]
- Dibner, C.; Schibler, U.; Albrecht, U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [PubMed]
- Shearman, L.P.; Sriram, S.; Weaver, D.R.; Maywood, E.S.; Chaves, I.; Zheng, B.; Kume, K.; Lee, C.C.; van der Horst, G.T.; Hastings, M.H.; et al. Interacting molecular loops in the mammalian circadian clock. Science 2000, 288, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 2001, 63, 647–676. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef] [PubMed]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the clock protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Hogenesch, J.B.; Gu, Y.Z.; Jain, S.; Bradfield, C.A. The basic-helix-loop-helix-pas orphan mop3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 1998, 95, 5474–5479. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Shearman, L.P.; Weaver, D.R.; Zylka, M.J.; de Vries, G.J.; Reppert, S.M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 1999, 96, 57–68. [Google Scholar] [CrossRef]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. Mcry1 and mcry2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef]
- Ramsey, K.M.; Marcheva, B.; Kohsaka, A.; Bass, J. The clockwork of metabolism. Annu. Rev. Nutr. 2007, 27, 219–240. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor rev-erbalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Triqueneaux, G.; Thenot, S.; Kakizawa, T.; Antoch, M.P.; Safi, R.; Takahashi, J.S.; Delaunay, F.; Laudet, V. The orphan receptor rev-erbalpha gene is a target of the circadian clock pacemaker. J. Mol. Endocrinol. 2004, 33, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Akashi, M.; Takumi, T. The orphan nuclear receptor roralpha regulates circadian transcription of the mammalian core-clock bmal1. Nat. Struct. Mol. Biol. 2005, 12, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Guillaumond, F.; Dardente, H.; Giguere, V.; Cermakian, N. Differential control of bmal1 circadian transcription by rev-erb and ror nuclear receptors. J. Biol. Rhythms 2005, 20, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Marcheva, B.; Ramsey, K.M.; Peek, C.B.; Affinati, A.; Maury, E.; Bass, J. Circadian clocks and metabolism. In Handbook of Experimental Pharmacology; Springer: Berlin, Germany, 2013; pp. 127–155. [Google Scholar]
- Eckel-Mahan, K.; Sassone-Corsi, P. Metabolism and the circadian clock converge. Physiol. Rev. 2013, 93, 107–135. [Google Scholar] [CrossRef] [PubMed]
- Opperhuizen, A.L.; van Kerkhof, L.W.; Proper, K.I.; Rodenburg, W.; Kalsbeek, A. Rodent models to study the metabolic effects of shiftwork in humans. Front. Pharmacol. 2015, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Purvis, T.E.; Mistretta, J.; Scheer, F.A. Effects of the internal circadian system and circadian misalignment on glucose tolerance in chronic shift workers. J. Clin. Endocrinol. Metab. 2016, 101, 1066–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheer, F.A.; Hilton, M.F.; Mantzoros, C.S.; Shea, S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 2009, 106, 4453–4458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arble, D.M.; Ramsey, K.M.; Bass, J.; Turek, F.W. Circadian disruption and metabolic disease: Findings from animal models. Best Pract. Res. Clin. Endocrinol. Metab. 2010, 24, 785–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barclay, J.L.; Husse, J.; Bode, B.; Naujokat, N.; Meyer-Kovac, J.; Schmid, S.M.; Lehnert, H.; Oster, H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS ONE 2012, 7, e37150. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokkan, K.A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Opperhuizen, A.L.; Wang, D.; Foppen, E.; Jansen, R.; Boudzovitch-Surovtseva, O.; de Vries, J.; Fliers, E.; Kalsbeek, A. Feeding during the resting phase causes profound changes in physiology and desynchronization between liver and muscle rhythms of rats. Eur. J. Neurosci. 2016, 44, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- De Goede, P.S.S.; Oosterman, J.E.; Foppen, E.; Jansen, R.; la Fleur, S.E.; Challet, E.; Kalsbeek, A. Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol. Sleep Circ. Rhythm. 2018, 4, 24–33. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Panda, S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 2017, 39, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Dyar, K.A.; Ciciliot, S.; Tagliazucchi, G.M.; Pallafacchina, G.; Tothova, J.; Argentini, C.; Agatea, L.; Abraham, R.; Ahdesmaki, M.; Forcato, M.; et al. The calcineurin-nfat pathway controls activity-dependent circadian gene expression in slow skeletal muscle. Mol. Metab. 2015, 4, 823–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Reznick, J.; Preston, E.; Wilks, D.L.; Beale, S.M.; Turner, N.; Cooney, G.J. Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim. Biophys. Acta 2013, 1832, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.; Strubbe, J.H. Circadian control of insulin secretion is independent of the temporal distribution of feeding. Physiol. Behav. 1998, 63, 553–558. [Google Scholar] [CrossRef]
- Su, Y.; Foppen, E.; Zhang, Z.; Fliers, E.; Kalsbeek, A. Effects of 6-meals-a-day feeding and 6-meals-a-day feeding combined with adrenalectomy on daily gene expression rhythms in rat epididymal white adipose tissue. Genes Cells Devot. Mol. Cell. Mech. 2016, 21, 6–24. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Delgado, R.; Angeles-Castellanos, M.; Saderi, N.; Buijs, R.M.; Escobar, C. Food intake during the normal activity phase prevents obesity and circadian desynchrony in a rat model of night work. Endocrinology 2010, 151, 1019–1029. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.J.; Kumar, V.; Panda, S. A camera-phone based study reveals erratic eating pattern and disrupted daily eating-fasting cycle among adults in india. PLoS ONE 2017, 12, e0172852. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.; Panda, S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 2015, 22, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Foppen, E.; Mansur Machado, F.S.; Fliers, E.; Kalsbeek, A. The role of the daily feeding rhythm in the regulation of the day/night rhythm in triglyceride secretion in rats. Chronobiol. Int. 2018, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Drevet, K.; Pevet, P.; Challet, E. Daily meal timing is not necessary for resetting the main circadian clock by calorie restriction. J. Neuroendocrinol. 2008, 20, 251–260. [Google Scholar] [CrossRef] [PubMed]
- La Fleur, S.E.; Kalsbeek, A.; Wortel, J.; Buijs, R.M. A suprachiasmatic nucleus generated rhythm in basal glucose concentrations. J. Neuroendocrinol. 1999, 11, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Raingard, H.; Dumont, S.; Kalsbeek, A.; Vuillez, P.; Challet, E. Ultradian feeding in mice not only affects the peripheral clock in the liver, but also the master clock in the brain. Chronobiol. Int. 2017, 34, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.; Pevet, P.; Challet, E. Circadian and photic regulation of clock and clock-controlled proteins in the suprachiasmatic nuclei of calorie-restricted mice. Eur. J. Neurosci. 2007, 25, 3691–3701. [Google Scholar] [CrossRef] [PubMed]
- Cailotto, C.; La Fleur, S.E.; Van Heijningen, C.; Wortel, J.; Kalsbeek, A.; Feenstra, M.; Pevet, P.; Buijs, R.M. The suprachiasmatic nucleus controls the daily variation of plasma glucose via the autonomic output to the liver: Are the clock genes involved? Eur. J. Neurosci. 2005, 22, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Tahara, Y.; Saito, K.; Ohnishi, N.; Kubo, Y.; Seo, Y.; Otsuka, M.; Fuse, Y.; Ohura, Y.; Hirao, A.; et al. Meal frequency patterns determine the phase of mouse peripheral circadian clocks. Sci. Rep. 2012, 2, 711. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Sasaki, H.; Ohtsu, T.; Shiraishi, T.; Tahara, Y.; Shibata, S. Feeding and adrenal entrainment stimuli are both necessary for normal circadian oscillation of peripheral clocks in mice housed under different photoperiods. Chronobiol. Int. 2015, 32, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.; Fliers, E.; Romijn, J.A.; La Fleur, S.E.; Wortel, J.; Bakker, O.; Endert, E.; Buijs, R.M. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology 2001, 142, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Oishi, K.; Konishi, M.; Murata, Y.; Itoh, N. Time-imposed daily restricted feeding induces rhythmic expression of fgf21 in white adipose tissue of mice. Biochem. Biophys. Res. Commun. 2011, 412, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Markan, K.R.; Naber, M.C.; Ameka, M.K.; Anderegg, M.D.; Mangelsdorf, D.J.; Kliewer, S.A.; Mohammadi, M.; Potthoff, M.J. Circulating fgf21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes 2014, 63, 4057–4063. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N. Hormone-like (endocrine) fgfs: Their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010, 342, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, S.; Arimoto, A.; Kuramoto, Y.; Kozako, T.; Honda, S.; Shimeno, H.; Soeda, S. Fasting promotes the expression of sirt1, an nad+ -dependent protein deacetylase, via activation of pparalpha in mice. Mol. Cell. Biochem. 2010, 339, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Lundasen, T.; Hunt, M.C.; Nilsson, L.M.; Sanyal, S.; Angelin, B.; Alexson, S.E.; Rudling, M. Pparalpha is a key regulator of hepatic fgf21. Biochem. Biophys. Res. Commun. 2007, 360, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garaulet, M.; Martinez-Nicolas, A.; Ruiz, J.R.; Konstabel, K.; Labayen, I.; Gonzalez-Gross, M.; Marcos, A.; Molnar, D.; Widhalm, K.; Casajus, J.A.; et al. Fragmentation of daily rhythms associates with obesity and cardiorespiratory fitness in adolescents: The helena study. Clin. Nutr. 2016, 36, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Corbalan-Tutau, M.D.; Madrid, J.A.; Ordovas, J.M.; Smith, C.E.; Nicolas, F.; Garaulet, M. Differences in daily rhythms of wrist temperature between obese and normal-weight women: Associations with metabolic syndrome features. Chronobiol. Int. 2011, 28, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Luik, A.I.; Zuurbier, L.A.; Hofman, A.; Van Someren, E.J.; Tiemeier, H. Stability and fragmentation of the activity rhythm across the sleep-wake cycle: The importance of age, lifestyle, and mental health. Chronobiol. Int. 2013, 30, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lopez, N.; Tarabra, E.; Toledo, M.; Garcia-Macia, M.; Sahu, S.; Coletto, L.; Batista-Gonzalez, A.; Barzilai, N.; Pessin, J.E.; Schwartz, G.J.; et al. System-wide benefits of intermeal fasting by autophagy. Cell Metab. 2017, 26, 856–871. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Panda, S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 2016, 23, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, A.; Kobiita, A.; Damara, M.; Misra, N.; Meziane, H.; Champy, M.F.; Chambon, P. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master scn clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, E6691–E6698. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.S.; Ratcliffe, W.F.; Grenett, M.H.; Brewer, R.A.; Gamble, K.L.; Young, M.E. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int. J. Obes. 2013, 37, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Sherman, H.; Genzer, Y.; Cohen, R.; Chapnik, N.; Madar, Z.; Froy, O. Timed high-fat diet resets circadian metabolism and prevents obesity. FASEB J. 2012, 26, 3493–3502. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Turek, F.W. Timing of meals: When is as critical as what and how much. American journal of physiology. Endocrinol. Metab. 2017, 312, E369–E380. [Google Scholar]
- Patterson, R.E.; Sears, D.D. Metabolic effects of intermittent fasting. Annu. Rev. Nutr. 2017, 37, 371–393. [Google Scholar] [CrossRef] [PubMed]
- Collier, R. Intermittent fasting: The science of going without. CMAJ Can. Med. Assoc. J. 2013, 185, E363–E364. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.E.; Laughlin, G.A.; LaCroix, A.Z.; Hartman, S.J.; Natarajan, L.; Senger, C.M.; Martinez, M.E.; Villasenor, A.; Sears, D.D.; Marinac, C.R.; et al. Intermittent fasting and human metabolic health. J. Acad. Nutr. Diet. 2015, 115, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Stockman, M.C.; Thomas, D.; Burke, J.; Apovian, C.M. Intermittent fasting: Is the wait worth the weight? Curr. Obes. Rep. 2018, 7, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Nas, A.; Mirza, N.; Hagele, F.; Kahlhofer, J.; Keller, J.; Rising, R.; Kufer, T.A.; Bosy-Westphal, A. Impact of breakfast skipping compared with dinner skipping on regulation of energy balance and metabolic risk. Am. J. Clin. Nutr. 2017, 105, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Ohkawara, K.; Cornier, M.A.; Kohrt, W.M.; Melanson, E.L. Effects of increased meal frequency on fat oxidation and perceived hunger. Obesity 2013, 21, 336–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munsters, M.J.; Saris, W.H. Effects of meal frequency on metabolic profiles and substrate partitioning in lean healthy males. PLoS ONE 2012, 7, e38632. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Okauchi, H.; Hashimoto, C.; Wada, N.; Oishi, K. Determination of reference genes that are independent of feeding rhythms for circadian studies of mouse metabolic tissues. Mol. Genet. Metab. 2017, 121, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Dardente, H.; Menet, J.S.; Challet, E.; Tournier, B.B.; Pevet, P.; Masson-Pevet, M. Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Brain Res. Mol. Brain Res. 2004, 124, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Tournier, B.B.; Menet, J.S.; Dardente, H.; Poirel, V.J.; Malan, A.; Masson-Pevet, M.; Pevet, P.; Vuillez, P. Photoperiod differentially regulates clock genes’ expression in the suprachiasmatic nucleus of syrian hamster. Neuroscience 2003, 118, 317–322. [Google Scholar] [CrossRef]







| Genes | Two Way ANOVA Table (p Values) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Liver | Muscle | BAT | |||||||
| Feeding | Time | Interaction | Feeding | Time | Interaction | Feeding | Time | Interaction | |
| Bmal1 | =0.008 | <0.001 | =0.057 | =0.758 | <0.001 | =0.830 | =0.199 | <0.001 | =0.002 |
| Cry1 | <0.001 | <0.001 | <0.001 | =0.345 | <0.001 | =0.637 | =0.003 | <0.001 | =0.074 |
| Cry 2 | =0.270 | =0.021 | =0.516 | =0.342 | =0.353 | =0.670 | =0.051 | =0.178 | =0.684 |
| Per1 | =0.765 | <0.001 | =0.178 | =0.466 | <0.001 | =0.563 | =0.117 | <0.001 | =0.991 |
| Per2 | =0.072 | <0.001 | =0.021 | =0.888 | <0.001 | =0.003 | =0.262 | <0.001 | =0.175 |
| Dbp | =0.141 | <0.001 | =0.105 | =0.969 | <0.001 | <0.001 | =0.002 | <0.001 | <0.001 |
| Rev-erbα | =0.122 | <0.001 | <0.001 | =0.583 | <0.001 | =0.049 | =0.360 | <0.001 | =0.865 |
| Ucp1 | =0.166 | =0.039 | =0.654 | ||||||
| Ucp3 | =0.299 | =0.002 | =0.110 | ||||||
| Pparα | =0.644 | =0.122 | =0.459 | =0.252 | =0.870 | =0.471 | |||
| Pgc-1α | =0.526 | =0.927 | =0.848 | =0.340 | =0.031 | =0.271 | =0.311 | =0.079 | =0.121 |
| Glut2 | =0.505 | <0.001 | =0.234 | ||||||
| Glut4 | =0.093 | =0.066 | =0.684 | =0.415 | =0.126 | =0.531 | |||
| Pdk4 | =0.063 | =0.004 | =0.082 | =0.161 | =0.023 | =0.115 | =0.248 | =0.027 | =0.348 |
| Hsl | =0.803 | =0.954 | =0.359 | =0.559 | =0.427 | =0.776 | =0.485 | =0.187 | =0.546 |
| Lpl | =0.624 | <0.001 | =0.495 | =0.077 | =0.144 | =0.965 | =0.065 | =0.010 | =0.252 |
| Srebp1c | =0.840 | =0.282 | =0.026 | =0.379 | =0.015 | =0.997 | =0.224 | =0.110 | =0.287 |
| Sirt1 | =0.194 | =0.718 | =0.318 | ||||||
| Sirt3 | =0.104 | =0.045 | =0.606 | =0.818 | =0.023 | =0.341 | |||
| Fas | =0.053 | =0.006 | <0.001 | =0.459 | =0.101 | =0.832 | =0.836 | =0.027 | =0.581 |
| Fgf21 | =0.098 | <0.001 | =0.181 | =0.298 | =0.024 | =0.281 | |||
| Genes | Ad libitum | 6-Meal | ||||
|---|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | p Value | ||
| Bmal1 | a | 0.12 | 0.007 | 0.11 | 0.01 | =0.767 |
| b | 0.12 | 0.01 | 0.13 | 0.02 | =0.719 | |
| c | 0.7 | 0.32 | 0.3 | 0.60 | =0.595 | |
| Cry1 | a | 0.21 | 0.01 | 0.22 | 0.01 | =0.351 |
| b | 0.08 | 0.01 | 0.07 | 0.02 | =0.788 | |
| c | 21.2 | 0.71 | 21.1 | 1.28 | =0.953 | |
| Cry2 | a | 0.10 | 0.01 | 0.12 | 0.01 | =0.335 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Per1 | a | 0.03 | 0.003 | 0.04 | 0.01 | =0.463 |
| b | 0.03 | 0.01 | -- | -- | ||
| c | 14.0 | 0.75 | -- | -- | ||
| Per2 | a | 0.09 | 0.01 | 0.09 | 0.01 | =0.899 |
| b | 0.06 | 0.01 | 0.03 | 0.01 | =0.074 | |
| c | 14.8 | 0.86 | 12.7 | 1.28 | =0.169 | |
| Dbp | a | 1.29 | 0.09 | 1.28 | 0.08 | =0.973 |
| b | 1.33 | 0.13 | 1.22 | 0.12 | =0.548 | |
| c | 12.1 α | 0.37 | 10.4 | 0.38 | =0.002 | |
| Rev-erbα | a | 0.54 | 0.04 | 0.51 | 0.04 | =0.658 |
| b | 0.50 | 0.06 | 0.42 | 0.06 | =0.340 | |
| c | 8.6 | 0.44 | 7.2 | 0.50 | =0.055 | |
| Ucp3 | a | 0.10 | 0.01 | 0.14 | 0.03 | =0.286 |
| b | 0.07 | 0.02 | 0.15 | 0.04 | =0.122 | |
| c | 3.3 α | 0.80 | 23.8 | 1.10 | <0.001 | |
| Pgc-1α | a | 0.25 | 0.02 | 0.27 | 0.01 | =0.355 |
| b | -- | -- | 0.07 | 0.02 | ||
| c | -- | -- | 16.4 | 1.19 | ||
| Glut4 | a | 6.33 | 0.27 | 7.04 | 0.33 | =0.112 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Pdk4 | a | 1.86 | 0.16 | 2.78 | 0.60 | =0.148 |
| b | 0.90 | 0.22 | 2.48 | 0.85 | =0.077 | |
| c | 3.8 α | 0.95 | 23.5 | 1.30 | <0.001 | |
| Hsl | a | 0.06 | 0.01 | 0.07 | 0.01 | =0.549 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Lpl | a | 7.80 | 0.24 | 8.72 | 0.42 | =0.065 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Srebp1c | a | 0.09 | 0.01 | 0.11 | 0.02 | =0.364 |
| b | 0.05 | 0.01 | -- | -- | ||
| c | 23.2 | 0.92 | -- | -- | ||
| Sirt3 | a | 0.58 | 0.02 | 0.63 | 0.02 | =0.096 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Fas | a | 0.17 | 0.02 | 0.20 | 0.03 | =0.451 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Genes | Ad libitum | 6-Meal | p Value | |||
|---|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | |||
| Bmal1 | a | 0.01 | 0.001 | 0.01 | 0.001 | =0.193 |
| b | 0.01 α | 0.001 | 0.002 | 0.001 | <0.001 | |
| c | 22.1 α | 0.57 | 0.0 | 1.41 | <0.001 | |
| Cry1 | a | 0.02 α | 0.01 | 0.01 | 0.001 | <0.001 |
| b | 0.01 | 0.001 | 0.01 | 0.001 | =0.608 | |
| c | 20.1 | 0.89 | 20.1 | 0.48 | =0.978 | |
| Cry2 | a | 0.01 α | 0.001 | 0.01 | 0.001 | =0.041 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Per1 | a | 0.004 | 0.001 | 0.003 | 0.001 | =0.113 |
| b | 0.001 | 0.001 | 0.001 | 0.001 | =0.890 | |
| c | 3.0 | 1.44 | 2.5 | 0.96 | =0.801 | |
| Per2 | a | 0.01 | 0.001 | 0.004 | 0.001 | =0.286 |
| b | 0.01 | 0.001 | 0.003 | 0.001 | =0.103 | |
| c | 14.3 | 1.12 | 14.8 | 0.86 | =0.732 | |
| Dbp | a | 0.13 | 0.01 | 0.09 | 0.10 | =0.741 |
| b | 0.11 α | 0.02 | 0.03 | 0.004 | <0.001 | |
| c | 9.5 α | 0.55 | 13.1 β | 0.46 | <0.001 | |
| Rev-erbα | a | 0.04 | 0.004 | 0.03 | 0.002 | =0.359 |
| b | 0.02 | 0.01 | 0.02 | 0.003 | =0.885 | |
| c | 10.3 | 1.22 | 11.1 | 0.62 | =0.553 | |
| Ucp1 | a | 9.80 | 0.96 | 8.24 | 0.50 | =0.159 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Pparα | a | 0.01 | 0.01 | 0.05 | 0.003 | =0.247 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Pgc-1α | a | 0.04 | 0.004 | 0.03 | 0.002 | =0.313 |
| b | 0.02 | 0.01 | -- | -- | ||
| c | 12.7 | 1.47 | -- | -- | ||
| Glut4 | a | 0.74 | 0.07 | 0.67 | 0.04 | =0.41 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Pdk4 | a | 0.66 | 0.07 | 0.77 | 0.06 | =0.250 |
| b | -- | -- | 0.27 | 0.08 | ||
| c | -- | -- | 16.5 | 1.13 | ||
| Hsl | a | 0.13 | 0.01 | 0.12 | 0.01 | =0.485 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Lpl | a | 2.05 | 0.20 | 1.63 | 0.10 | =0.064 |
| b | 0.80 | 0.28 | -- | -- | ||
| c | 14.4 | 1.35 | -- | -- | ||
| Srebp1c | a | 0.05 | 0.01 | 0.04 | 0.004 | =0.219 |
| b | 0.02 | 0.01 | -- | -- | ||
| c | 17.1 | 1.44 | -- | -- | ||
| Sirt3 | a | 0.12 | 0.004 | 0.12 | 0.003 | =0.826 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Fas | a | 5.75 | 0.29 | 5.67 | 0.18 | =0.833 |
| b | -- | -- | 0.85 | 0.26 | ||
| c | -- | -- | 14.5 | 1.15 | ||
| Fgf21 | a | 0.01 | 0.01 | 0.01 | 0.001 | =0.352 |
| b | -- | -- | 0.001 | 0.001 | ||
| c | -- | -- | 8.1 | 1.28 | ||
| Genes | Ad libitum | 6-Meal | p Value | |||
|---|---|---|---|---|---|---|
| Mean | SEM | Mean | SEM | |||
| Bmal1 | a | 0.04 α | 0.003 | 0.03 | 0.003 | <0.001 |
| b | 0.40 α | 0.01 | 0.02 | 0.003 | =0.015 | |
| c | 1.0 | 0.50 | 1.5 | 0.52 | =0.402 | |
| Cry1 | a | 0.05 α | 0.001 | 0.03 | 0.002 | <0.001 |
| b | 0.03 α | 0.002 | 0.02 | 0.003 | <0.001 | |
| c | 21.3 | 0.26 | 21.7 | 0.62 | =0.505 | |
| Cry2 | a | 0.09 | 0.01 | 0.08 | 0.01 | =0.271 |
| b | 0.03 | 0.01 | -- | -- | ||
| c | 20.3 | 1.40 | -- | -- | ||
| Per1 | a | 0.03 | 0.003 | 0.04 | 0.004 | =0.762 |
| b | 0.03 | 0.004 | 0.01 | 0.01 | =0.092 | |
| c | 15.2 | 0.62 | 14.1 | 1.25 | =0.449 | |
| Per2 | a | 0.10 | 0.01 | 0.08 | 0.01 | =0.070 |
| b | 0.07 α | 0.01 | 0.04 | 0.01 | <0.001 | |
| c | 17.6 | 0.50 | 17.0 | 0.95 | =0.602 | |
| Dbp | a | 0.23 | 0.03 | 0.18 | 0.01 | =0.158 |
| b | 0.30 | 0.04 | 0.20 | 0.02 | =0.087 | |
| c | 12.0 | 0.56 | 13.1 | 0.40 | =0.100 | |
| Rev-erbα | a | 0.19 | 0.03 | 0.15 | 0.01 | =0.184 |
| b | 0.25 α | 0.04 | 0.16 | 0.01 | =0.021 | |
| c | 9.4 α | 0.56 | 11.2 | 0.32 | =0.005 | |
| Pparα | a | 0.18 | 0.01 | 0.19 | 0.01 | =0.636 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Pgc-1α | a | 0.01 | 0.001 | 0.01 | 0.001 | =0.444 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Glut2 | a | 0.39 | 0.02 | 0.40 | 0.02 | =0.512 |
| b | 0.09 | 0.02 | 0.09 | 0.03 | =0.951 | |
| c | 14.1 | 1.30 | 13.1 | 1.07 | =0.533 | |
| Pdk4 | a | 0.09 | 0.01 | 0.07 | 0.01 | =0.056 |
| b | 0.04 | 0.01 | -- | -- | ||
| c | 3.9 | 0.96 | -- | -- | ||
| Hsl | a | 0.01 | 0.001 | 0.01 | 0.001 | =0.815 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Lpl | a | 0.01 | 0.001 | 0.01 | 0.001 | =0.646 |
| b | -- | -- | 0.004 | 0.001 | ||
| c | -- | -- | 7.2 | 0.80 | ||
| Srebp1c | a | 0.64 | 0.05 | 0.63 | 0.05 | =0.841 |
| b | -- | -- | 0.22 | 0.07 | ||
| c | -- | -- | 7.9 | 1.10 | ||
| Sirt1 | a | 0.02 | 0.002 | 0.02 | 0.001 | =0.178 |
| b | -- | -- | -- | -- | ||
| c | -- | -- | -- | -- | ||
| Fas | a | 1.08 | 0.08 | 1.31 | 0.10 | =0.068 |
| b | 0.40 | 0.11 | 0.53 | 0.13 | =0.482 | |
| c | 17.7 α | 1.13 | 9.2 | 0.97 | <0.001 | |
| Fgf21 | a | 0.04 | 0.003 | 0.03 | 0.003 | =0.103 |
| b | 0.01 | 0.004 | -- | -- | ||
| c | 6.4 | 0.99 | -- | -- | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Goede, P.; Sen, S.; Su, Y.; Foppen, E.; Poirel, V.-J.; Challet, E.; Kalsbeek, A. An Ultradian Feeding Schedule in Rats Affects Metabolic Gene Expression in Liver, Brown Adipose Tissue and Skeletal Muscle with Only Mild Effects on Circadian Clocks. Int. J. Mol. Sci. 2018, 19, 3171. https://doi.org/10.3390/ijms19103171
De Goede P, Sen S, Su Y, Foppen E, Poirel V-J, Challet E, Kalsbeek A. An Ultradian Feeding Schedule in Rats Affects Metabolic Gene Expression in Liver, Brown Adipose Tissue and Skeletal Muscle with Only Mild Effects on Circadian Clocks. International Journal of Molecular Sciences. 2018; 19(10):3171. https://doi.org/10.3390/ijms19103171
Chicago/Turabian StyleDe Goede, Paul, Satish Sen, Yan Su, Ewout Foppen, Vincent-Joseph Poirel, Etienne Challet, and Andries Kalsbeek. 2018. "An Ultradian Feeding Schedule in Rats Affects Metabolic Gene Expression in Liver, Brown Adipose Tissue and Skeletal Muscle with Only Mild Effects on Circadian Clocks" International Journal of Molecular Sciences 19, no. 10: 3171. https://doi.org/10.3390/ijms19103171
APA StyleDe Goede, P., Sen, S., Su, Y., Foppen, E., Poirel, V.-J., Challet, E., & Kalsbeek, A. (2018). An Ultradian Feeding Schedule in Rats Affects Metabolic Gene Expression in Liver, Brown Adipose Tissue and Skeletal Muscle with Only Mild Effects on Circadian Clocks. International Journal of Molecular Sciences, 19(10), 3171. https://doi.org/10.3390/ijms19103171