Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle
Abstract
:1. Introduction
2. Results
2.1. Deletion of Bmal1 Gene Has No Effects on the Body Weight, Behavior, or Muscular Structure
2.2. Deletion of Bmal1 Gene in the Muscle Increases Muscular Oxidative Capacity
2.3. Deletion of the Bmal1 Gene in the Muscle Improves the State of Diet-Induced Obesity
2.4. Deletion of the Bmal1 Gene in the Muscle Improves the State of Insulin Sensitivity in Obesity
2.5. Deletion of the Bmal1 Gene in the Muscle Induces Gene Expression Related to Lipid Metabolism
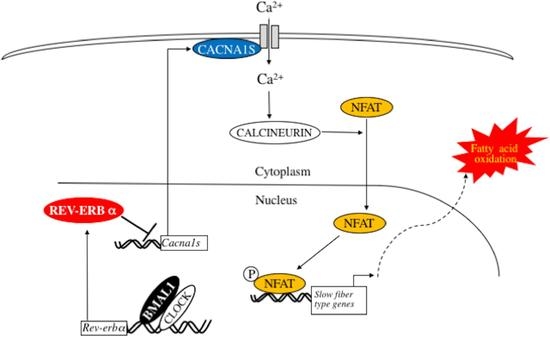
2.6. Deletion of Bmal1 Gene Activates Calcium Signaling in the Skeletal Muscle
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Locomotor Activity Rhythm Analysis
4.3. Metabolic Studies
4.4. Biochemical Analysis of Blood and Tissue
4.5. Gene Expression (Quantitative Reverse Transcription Polymerase Chain Reaction [qRT-PCR])
4.6. Exercise Performance Test
4.7. Histological Analyses
4.8. Preparation of Tissue Extract
4.9. Measurement of 3-Hydroxyacyl CoA Dehydrogenase Activity
4.10. Electron Microscopy
4.11. Western Blot Analysis
4.12. Plasmid Construction, Cell Culture and Cell Transfection
4.13. Chromatin Immunoprecipitation (ChIP) Assays
- Cacna1s −430; 5′-AGAAACTTAATCTCCATCTAAGG-3′,
- Cacna1s −282; 5′-GTGACTTATTATATCCAGGCTTG-3′,
- Cacna1s −2924; 5′-TCCTCTGTAAAAAGCAGTACCTGC-3′,
- Cacna1s −2774; 5′-CAGCCATTAATATCATTTCCTCTG-3′,
- Bmal1 −53; 5′-GGAAAGTAGGTTAGTGGTGCGAC-3′,
- and Bmal1 +31; 5′-AAGTCCGGCGCGGGTAAACAGG-3′.
4.14. Statistical Analysis
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Acetyl-CoA carboxylase |
| AKT | Protein kinase B |
| AMPK | AMP-activated protein kinase. |
| BMAL | Brain and muscle arnt-like protein 1 |
| Cd | Cluster of differentiation |
| Cpt | Carnitine palmitoyl transferase |
| Dgat1 | Diacylglycerol O-acyltransferase 1 |
| EDL | Extensor digitorum longus |
| Erra | Estrogen-related receptor-α |
| Fatp | Fatty acid transporter |
| Fasn | Fatty acid synthase |
| Flk | Fetal liver kinase |
| Glut4 | Glucose transporter type 4 |
| GN | Gastrocnemius |
| Hadh | Hydroxyacyl CoA dehydrogenase |
| HFD | High fat diet |
| Il | Interleukin |
| Irs | Insulin receptor substrate |
| Lcad | Long-chain acyl-CoA dehydrogenase |
| Mb | Myoglobin |
| Mcad | Medium-chain acyl-CoA dehydrogenase |
| Mcp1 | Monocyte chemotactic protein-1 |
| MyHC | Myosin heavy chain isoform |
| NFAT | Nuclear Factor of Activated T cells |
| Pgc-1 | Peroxisome proliferator-activated receptor gammer coactivator-1 |
| Ppar | Peroxisome proliferator-activated receptor |
| Rev-erb | Reverse orientation the c-erbA-1 gene |
| RORE | Retinoic acid receptor-related orphan receptor-responsive element |
| Scad | Short-chain acyl-CoA dehydrogenase |
| Scd1 | Stearoyl-coA desaturase 1 |
| Sol | Soleus |
| Tie2 | TEK receptor tyrosine kinase 2 |
| Tfam | Mitochondrial transcription factor A |
| Tnf | Tumor necrosis factor |
| Tnni1 | Troponin I solow |
| Tnni2 | Troponin I fast |
| Vlcad | Very long-chain acyl-CoA dehydrogenase |
| WAT | White adipose tissue |
| ZT | Zeitgeber time |
References
- Ikeda, M.; Nomura, M. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem. Biophys. Res. Commun. 1997, 233, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Hogenesch, J.B.; Gu, Y.Z.; Jain, S.; Bradfield, C.A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 1988, 95, 5474–5479. [Google Scholar] [CrossRef]
- Takahata, S.; Sogawa, K.; Kobayashi, A.; Ema, M.; Mimura, J.; Ozaki, N.; Fujii-Kuriyama, Y. Transcriptionally active heterodimer formation of an Arnt-like PAS protein, Arnt3, with HIF-1a, HLF and clock. Biochem. Biophys. Res. Commun. 1998, 248, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the clock protein in the mammalian circadian mechanism. Science 1988, 280, 1564–1569. [Google Scholar] [CrossRef]
- Yoo, S.H.; Ko, C.H.; Lowrey, P.L.; Buhr, E.D.; Song, E.J.; Chang, S.; Yoo, O.J.; Yamazaki, S.; Lee, C.; Takahashi, J.S. A noncanonical E-box enhancer drives mouse period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 2608–2613. [Google Scholar] [CrossRef] [PubMed]
- Kumaki, Y.; Ukai-Tadenuma, M.; Uno, K.D.; Nishio, J.; Masumoto, K.H.; Nagano, M.; Komori, T.; Shigeyoshi, Y.; Hogenesch, J.B.; Ueda, H.R. Analysis and synthesis of high-amplitude cis-elements in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 14946–14951. [Google Scholar] [CrossRef] [PubMed]
- Bunger, M.K.; Walisser, J.A.; Sullivan, R.; Manley, P.A.; Moran, S.M.; Kalscheur, V.L.; Colman, R.J.; Bradfield, C.A. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis 2005, 41, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, J.L.; Zhang, X.; McCarthy, J.J.; McDearmon, E.L.; Hornberger, T.A.; Russell, B.; Campbell, K.S.; Arbogast, S.; Reid, M.B.; Walker, J.R.; et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Nat. Acad. Sci. USA 2010, 107, 19090–19095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimba, S.; Ogawa, T.; Hitosugi, S.; Ichihashi, Y.; Nakadaira, Y.; Kobayashi, M.; Tezuka, M.; Kosuge, Y.; Ishige, K.; Ito, Y.; et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE 2011, 6, e25231. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, F.; Matsubara, C.; Myung, J.; Yoritaka, T.; Kamimura, N.; Tsutsumi, S.; Kanai, A.; Suzuki, Y.; Sassone-Corsi, P.; Aburatani, H.; et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol. Cell. Biol. 2010, 30, 5636–5648. [Google Scholar] [CrossRef] [PubMed]
- Rey, G.; Cesbron, F.; Rougemont, J.; Reinke, H.; Brunner, M.; Naef, F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011, 9, e1000595. [Google Scholar] [CrossRef] [PubMed]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Yoshitane, H.; Ozaki, H.; Terajima, H.; Du, N.H.; Suzuki, Y.; Fujimori, T.; Kosaka, N.; Shimba, S.; Sugano, S.; Takagi, T.; et al. CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol. Cell. Biol. 2014, 34, 1776–1787. [Google Scholar] [CrossRef] [PubMed]
- McDearmon, E.L.; Patel, K.N.; Ko, C.H.; Walisser, J.A.; Schook, A.C.; Chong, J.L.; Wilsbacher, L.D.; Song, E.J.; Hong, H.K.; Bradfield, C.A.; et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 2006, 314, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Zambon, A.C.; McDearmon, E.L.; Salomonis, N.; Vranizan, K.M.; Johansen, K.L.; Adey, D.; Takahashi, J.S.; Schambelan, M.; Conklin, B.R. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003, 4, R61. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.J.; Andrews, J.L.; McDearmon, E.L.; Campbell, K.S.; Barber, B.K.; Miller, B.H.; Walker, J.R.; Hogenesch, J.B.; Takahashi, J.S.; Esser, K.A. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genom. 2007, 31, 86–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, A.M.; Elliott, J.A.; Duffy, P.; Blake, C.M.; Ben Attia, S.; Katz, L.M.; Browne, J.A.; Gath, V.; McGivney, B.A.; Hill, E.W.; et al. Circadian regulation of locomotor activity and skeletal muscle gene expression in the horse. J. Appl. Physiol. 2010, 109, 1328–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyar, K.A.; Ciciliot, S.; Wright, L.E.; Biensø, R.S.; Tagliazucchi, G.M.; Patel, V.R.; Forcato, M.; Paz, M.I.; Gudiksen, A.; Solagna, F.; et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 2014, 3, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Harfmann, B.D.; Schroder, E.A.; Kachman, M.T.; Hodge, B.A.; Zhang, X.; Esser, K.A. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet. Muscle 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Schroder, E.A.; Harfmann, B.D.; Zhang, X.; Srikuea, R.; England, J.H.; Hodge, B.A.; Wen, Y.; Riley, L.A.; Yu, Q.; Christie, A.; et al. Intrinsic muscle clock is necessary for musculoskeletal health. J. Physiol. 2015, 593, 5387–5404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olefsky, J.M.; Glass, C.K. Macrophages, inflammation and insulin resistance. Annu. Rev. Physiol. 2010, 72, 219–246. [Google Scholar] [CrossRef] [PubMed]
- Arita, Y.; Kihara, S.; Ouchi, N.; Takahashi, M.; Maeda, K.; Miyagawa, J.; Hotta, K.; Shimomura, I.; Nakamura, T.; Miyaoka, K.; et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999, 257, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Hotta, K.; Funahashi, T.; Arita, Y.; Takahashi, M.; Matsuda, M.; Okamoto, Y.; Iwahashi, H.; Kuriyama, H.; Ouchi, N.; Maeda, K.; et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1595–1599. [Google Scholar] [CrossRef] [PubMed]
- Schulz, R.A.; Yutzey, K.E. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 2004, 266, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Okamura, H.; Aramburu, J.; García-Rodríguez, C.; Viola, J.P.; Raghavan, A.; Tahiliani, M.; Zhang, X.; Qin, J.; Hogan, P.G.; Rao, A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 2000, 6, 539–550. [Google Scholar] [CrossRef]
- Zhao, M.; New, L.; Kravchenko, V.V.; Kato, Y.; Gram, H.; di Padova, F.; Olson, E.N.; Ulevitch, R.J.; Han, J. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 1999, 19, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Michael, L.F.; Wu, Z.; Cheatham, R.B.; Puigserver, P.; Adelmant, G.; Lehman, J.J.; Kelly, D.P.; Spiegelman, B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 2001, 98, 3820–3825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Moorsel, D.; Hansen, J.; Havekes, B.; Scheer, F.A.; Jörgensen, J.A.; Hoeks, J.; Schrauwen-Hinderling, V.B.; Duez, H.; Lefebvre, P.; Schaper, N.C.; et al. Demonstration of a day-night rhythm in human skeletal muscle oxidative capacity. Mol. Metab. 2016, 5, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Naya, F.J.; Mercer, B.; Shelton, J.; Richardson, J.A.; Williams, R.S.; Olson, E.N. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 2000, 275, 4545–4548. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kanatous, S.B.; Thurmond, F.A.; Gallardo, T.; Isotani, E.; Bassel-Duby, R.; Williams, R.S. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 2002, 296, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Piétri-Rouxel, F.; Gentil, C.; Vassilopoulos, S.; Baas, D.; Mouisel, E.; Ferry, A.; Vignaud, A.; Hourdé, C.; Marty, I.; Schaeffer, L.; et al. DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 2010, 29, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Pircher, P.; Chomez, P.; Yu, F.; Vennstrom, B.; Larsson, L. Aberrant expression of myosin isoforms in skeletal muscles from mice lacking the rev-erbA α orphan receptor gene. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R482–R490. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Storch, K.F.; Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 2008, 105, 15172–15177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcheva, B.; Ramsey, K.M.; Buhr, E.D.; Kobayashi, Y.; Su, H.; Ko, C.H.; Ivanova, G.; Omura, C.; Mo, S.; Vitaterna, M.H.; et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010, 466, 627–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, B.; Knutsson, A.; Lindahl, B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup. Environ. Med. 2001, 58, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Ruger, M.; Scheer, F.A. Effects of circadian disruption on the cardiometabolic system. Rev. Endocr. Metab. Disord. 2009, 10, 245–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, Y.; Yang, C.; Tong, X.; Sun, H.; Cong, Y.; Yin, X.; Li, L.; Cao, S.; Dong, X.; Gong, Y.; et al. Shift work and diabetes mellitus: A meta-analysis of observational studies. Occup. Environ. Med. 2015, 72, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Buchvold, H.V.; Pallesen, S.; Oyane, N.M.; Bjorvatn, B. Associations between night work and BMI, alcohol, smoking, caffeine and exercise a cross-sectional study. BMC Pub. Health 2015, 15, 1112. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, R.W.; Feeback, D.L. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fibre types I, IIA, IIB and IIC. Stain Technol. 1990, 65, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Seligman, A.M.; Karnovsky, M.J.; Wasserkrug, H.L.; Hanker, J.S. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB). J. Cell Biol. 1968, 38, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.; Brdiczca, D.; Eyer, P.; Hofer, S.; Pette, D. Metabolic differentiation of distinct muscle types at the level of enzymatic organization. Eur. J. Biochem. 1969, 10, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Sunaga, H.; Ohkawara, R.; Shimba, S. Aryl hydrocarbon receptor modulates NADPH oxidase activity via direct transcriptional regulation of p40phox expression. Mol. Pharmacol. 2013, 83, 1133–1140. [Google Scholar] [CrossRef] [PubMed]










| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| 36B4 | GCACTTTCGCTTTCTGGAGGGTGTC | TGACTTGGTGCTTTGGCGGGATTAG |
| Cacna1s | CCACACAGGGTAGCATGTAA | TCTCAGCTCCTCGTTAGCTT |
| Cd36 | TGCTGGAGGTGTTATTGGTG | TGGGTTTTGCACATCAAAGA |
| Cd68 | CTTCCCACAGGCAGCACAG | AATGATGAGAGGCAGCAAGAGG |
| Cpt1b | GTCGCTTCTTCAACGTCTGG | AAGAAAGCAGCACGTTCGAT |
| Cpt2 | TCCTCGATCAAGATGGGAAC | GATCCTTCATCGGGAAGTCA |
| Dgat1 | TGTGTGGTGATGCTGATCCTGAGT | GCCAGGCGCTTCTCAATCTGAAAT |
| Erra | GGAGGACGGCAGAAGTACAAA | GCGACACCAGAGCGTTCAC |
| F4/80 | GTTTGGCTATGGGCTTCCAGTC | GCAAGGAGGACAGAGTTTATGGTG |
| Fasn | TGCTCCCAGCTGCAGGC | GCCCGGTAGCTCTGGGTGTA |
| Fatp1 | TGGTCAAGGTCAATGAGGACACGA | ACGCTGTGGGCAATCTTCTTGTTG |
| Fatp4 | TAGCCGCATCCTGTCCTTTGTGTA | CTTCTTGTTGTTGGCACCCTGGTT |
| Flk | CCCGCATGAAATTGAGCTAT | AAACATCTTCGCCACAGTCC |
| Glut4 | GCTTTGTGGCCTTCTTTGAG | CGGCAAATAGAAGGAAGACG |
| Hadha | TGTGCCTGCTGCATTTGACA | ACAAGGCCTTTGCTCTGCTT |
| Hadhb | AGCTGCACTTTCGGGTTTGT | AACAGCTGTGGTCATGGCTT |
| Il-1b | CAACCAACAAGTGATATTCTCCATG | GATCCACACTCTCCAGCTGCA |
| Il-6 | ACAACCACGGCCTTCCCTACT | CACGATTTCCCAGAGAACATG |
| Il-10 | ACTTGGGTTGCCAAGCCTTA | AGAAATCGATGACAGCGCCT |
| Insulin receptor | AAAGTTTGCCCAACCATCTG | GTGAAGGTCTTGGCAGAAGC |
| Irs1 | GCTCTAGTGCTTCCGTGTCC | GTTGCCACCCCTAGACAAAA |
| Lcad | ATGGCAAAATACTGGGCATC | TCTTGCGATCAGCTCTTTCA |
| Mb | CCTGGGTACCATCCTGAAGA | GAGCATCTGCTCCAAAGTCC |
| Mcad | CGCTCTTAGGACTACTTGCTAACC | ATGGTATTTACATGCAATGGACAG |
| Mcp1 | CTTCTGGGCCTGCTGTTCA | CCAGCCTACTCATTGGGATCA |
| MyHC I | GCCTGGGCTTACCTCTCTATCAC | CTTCTCAGACTTCCGCAGGAA |
| MyHC IIa | CAGCTGCACCTTCTCGTTTG | CCCGAAAACGGCCATCT |
| MyHC IIb | CAATCAGGAACCTTCGGAACAC | GTCCTGGCCTCTGAGAGCAT |
| MyHC IIx | GGACCCACGGTCGAAGTTG | CCCGAAAACGGCCATCT |
| Pgc1a | GATGGCACGCAGCCCTAT | CTCGCACGGAGAGTTAAAGGAA |
| Pgc1b | AACCCAACCAGTCTCACACAGG | ATGCTGTCCTTGTGGGTAGG |
| Ppara | ATGCCAGTACTGCCCTTTTC | GGCCTTGACCTTGTTCATGT |
| Scad | CCACCAGACAAGACCGATTT | TCAATGAGGTATGGCACC |
| Scd1 | TGGGTTGGCTGCTTGTG | GCGTGGGCAGGATGAAG |
| Tfam | CAAGTCAGCTGATGGGTATGG | TTTCCCTGAGCCGAATCATCC |
| Tie2 | TTGAAGTGACGAATGAGAT | ATTTAGAGCTGTCTGGCTT |
| Tnfa | CGTCAGCCGATTTGCTATCT | CGGACTCCGCAAAGTCTAAG |
| Tnni1 | TCATGCTGAAGAGCCTGATG | GGAGGCATTTGGCTTCAATA |
| Tnni2 | CTGAGGGGCAAGTTCAATA | AGGTCCCGTTCCTTCTCAGT |
| Vlcad | TCATTGCCAAGGGCGGTTGAT | TTTGCTGATGGCGGCTTCTA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wada, T.; Ichihashi, Y.; Suzuki, E.; Kosuge, Y.; Ishige, K.; Uchiyama, T.; Makishima, M.; Nakao, R.; Oishi, K.; Shimba, S. Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle. Int. J. Mol. Sci. 2018, 19, 2813. https://doi.org/10.3390/ijms19092813
Wada T, Ichihashi Y, Suzuki E, Kosuge Y, Ishige K, Uchiyama T, Makishima M, Nakao R, Oishi K, Shimba S. Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle. International Journal of Molecular Sciences. 2018; 19(9):2813. https://doi.org/10.3390/ijms19092813
Chicago/Turabian StyleWada, Taira, Yuya Ichihashi, Emi Suzuki, Yasuhiro Kosuge, Kumiko Ishige, Taketo Uchiyama, Makoto Makishima, Reiko Nakao, Katsutaka Oishi, and Shigeki Shimba. 2018. "Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle" International Journal of Molecular Sciences 19, no. 9: 2813. https://doi.org/10.3390/ijms19092813
APA StyleWada, T., Ichihashi, Y., Suzuki, E., Kosuge, Y., Ishige, K., Uchiyama, T., Makishima, M., Nakao, R., Oishi, K., & Shimba, S. (2018). Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle. International Journal of Molecular Sciences, 19(9), 2813. https://doi.org/10.3390/ijms19092813