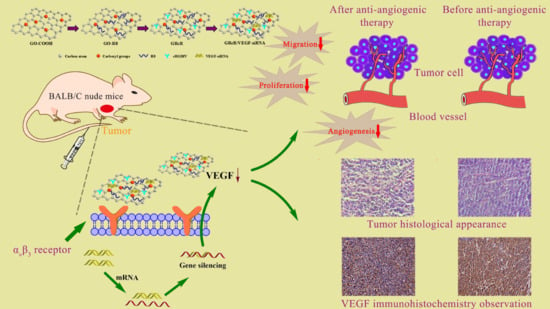
Preparation and Characterization of Functionalized Graphene Oxide Carrier for siRNA Delivery
Abstract
:1. Introduction
2. Results
2.1. Characterization (Fourier Transform Infrared Spectroscopy (FTIR), Ultraviolet Visible Spectrophotometer (UV), and Transmission Electron Microscope (TEM))
2.2. Tyndall Effect, Zeta Potential, Stability, and Particle Size Studies
2.3. Differential Scanning Calorimeter (DSC) and Thermal Gravimetric Analyzer (TGA) Spectra
2.4. Gel Retardation Assay
2.5. Release Profile of VEGF-siRNA
2.6. Cytotoxicity Assay
2.7. Cellular Uptake of GRcR-VEGF-siRNA
2.8. Tumor Growth Inhibition In Vitro
2.9. Gene Silencing Effect Evaluation on mRNA Level
2.10. Gene Silencing Effect Evaluation on Protein Level
2.11. Tumors Inhibition Assay In Vivo
2.12. Distribution of GRcR/VEGF-siRNA In Vivo
2.13. Inhibition of VEGF-Induced Angiogenesis
2.14. Histological and Immunohistochemical Study of Tumor Tissue
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Carboxylation of Nano-Graphene Oxide Sheets (NGOS–COOH)
4.3. Preparation of GRcR Complex
4.4. Gel Retardation Assay and Preparation of GRcR/VEGF-siRNA
4.5. Release Profile of GRcR/VEGF-siRNA
4.6. Cell Culture
4.7. Cytotoxicity Assay
4.8. Cellular Uptake of GRcR/VEGF-siRNA
4.9. Antiproliferation Assay
4.10. Real Time PCR
4.11. ELISA
4.12. Tumor Growth Inhibition of GRcR/VEGF-siRNA In Vivo
4.13. Tumor Targeting Ability of GRcR/VEGF-siRNA
4.14. Matrigelangiogenesis Assay
4.15. Histological and Immunohistochemical Observation
4.16. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bodak, M.; Cirera-Salinas, D.; Luitz, J.; Ciaudo, C. The Role of RNA interference in stem cell biology: Beyond the mutant phenotypes. J. Mol. Biol. 2017, 429, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Pena, C.; Kamen, A.A. RNA interference technology to improve the baculovirus-insect cell expression system. Biotechnol. Adv. 2018, 36, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Artigas, F.; Celada, P.; Bortolozzi, A. Can we increase the speed and efficacy of antidepressanttreatments? Part II. Glutamatergic and RNA interference strategies. Eur. Neuropsychopharmacol. 2018, 28, 457–482. [Google Scholar] [CrossRef] [PubMed]
- Lozada-Delgado, E.L.; Grafals-Ruiz, N.; Vivas-Mejía, P.E. RNA interference for glioblastoma therapy: Innovation ladder from the bench to clinical trials. Life Sci. 2017, 188, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Srimanee, A.; Arvanitidou, M.; Kim, K.; Hällbrink, M.; Langel, Ü. Cell-penetrating peptides for siRNA delivery to glioblastomas. Peptides 2018, 104, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Zahir, F.; Mottaghitalab, F.; Dinarvand, M.; Atyabi, F. siRNA delivery for treatment of degenerative diseases, new hopes and challenges. J. Drug Deliv. Sci. Technol. 2018, 45, 428–441. [Google Scholar] [CrossRef]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Barba, A.A.; Cascone, S.; Caccavo, D.; Lamberti, G.; Chiarappa, G.; Abrami, M.; Grassi, G.; Grassi, M.; Tomaiuolo, G.; Guido, S.; et al. Engineering approaches in siRNA delivery. Int. J. Pharm. 2018, 1860, 343–358. [Google Scholar] [CrossRef] [PubMed]
- McCallion, C.; Burthem, J.; Rees-Unwin, K.; Golovanov, A.; Pluen, A. Graphene in therapeutics delivery: Problems, solutions and future opportunities. Eur. J. Pharm. Biopharm. 2016, 104, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, H.; Zhao, C.; Qin, G.; Xi, G.; Li, T.; Wang, X.; Chen, T. One-step reduction and PEGylation of graphene oxide for photothermally controlled drug delivery. Biomaterials 2014, 35, 4986–4995. [Google Scholar] [CrossRef] [PubMed]
- Justin, R.; Chen, B. Characterisation and drug release performance of biodegradablechitosan–graphene oxide nanocomposites. Carbohydr. Polym. 2014, 103, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Shim, G.; Kim, J.Y.; Han, J.; Chung, S.W.; Lee, S.; Byun, Y.; Oh, Y.K. Reduced graphene oxide nanosheets coated with an anti-angiogenic anticancer low-molecular-weight heparin derivative for delivery of anticancer drugs. J. Control. Release 2014, 189, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhou, L.; Wei, S.; Ge, X.; Zhou, J.; Jiang, H.; Li, F.; Shen, J. Combination of chemotherapy and photodynamic therapy using graphene oxide as drug delivery system. J. Photochem. Photobiol. B 2014, 135, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.K.; Goyal, R.; Gupta, K.C.; Kumar, P. Functionalized graphene oxide mediated nucleic acid delivery. Carbon 2013, 51, 224–235. [Google Scholar] [CrossRef]
- Feng, L.; Yang, X.; Shi, X.; Tan, X.; Peng, R.; Wang, J.; Liu, Z. Polyethylene glycol and polyethylenimine dual-functionalized nano-graphene oxide for photothermally enhanced gene delivery. Small 2013, 9, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Niu, G.L.; Cao, X.F.; Wen, Y.K.; Xiang, R.; Duan, H.J.; Chen, Y.S. The preparation of functionalized graphene oxide for targeted intracellular delivery of siRNA. J. Mater. Chem. 2012, 22, 6649–6654. [Google Scholar] [CrossRef]
- Yukawa, H.; Kagami, Y.; Watanabe, M.; Oishi, K.; Miyamoto, Y.; Okamoto, Y.; Tokeshi, M.; Kaji, N.; Noguchi, H.; Ono, K.; et al. Quantum dots labeling using octa-arginine peptides for imaging of adipose tissue-derived stem cells. Biomaterials 2010, 31, 4094–4103. [Google Scholar] [CrossRef] [PubMed]
- Kamei, N.; Bech Nielsen, E.J.; Nakakubo, T.; Aoyama, Y.; Rahbek, U.L.; Pedersen, B.L.; Takeda-Morishita, M. Applicability and limitations of cell-penetrating peptides innoncovalent mucosal drug or carrier delivery systems. J. Pharmaceut. Sci. 2016, 105, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Golan, M.; Feinshtein, V.; David, A. Conjugates of HA2 with octaarginine-grafted HPMA copolymer offer effective siRNA delivery and gene silencing in cancer cells. Eur. J. Pharm. Biopharm. 2016, 109, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; He, B.; Jin, H.; Zhang, H.; Dai, W.; Zhang, L.; Wang, X.; Wang, J.; Zhang, X.; Zhang, Q. Targeting efficiency of RGD-modified nanocarriers with different ligand intervals in response to integrin αvβ3 clustering. Biomaterials 2014, 35, 6106–6117. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Li, J.; Cao, Z.; Yang, P.; Qiu, Y.; Yang, B.; Wang, Y.; Long, Y.; Liu, Y.; Zhang, Q.; et al. A pH-responsive cell-penetrating peptide-modified liposomes with active recognizing of integrin αvβ3 for the treatment of melanoma. J. Control. Release 2015, 217, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, B.; Zou, M.; Cheng, G. Cyclic RGD-modified chitosan/graphene oxide polymers for drug delivery and cellular imaging. Colloids Surf. B 2014, 122, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Chen, B.; Zou, M. In vivo pharmacokinetics, biodistribution and the anti-tumor effect of cyclic RGD-modified doxorubicin-loaded polymers in tumor-bearing mice. Colloids Surf. B 2016, 146, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Allman, R.; Cowburn, P.; Mason, M. In vitro and in vivo effects of a cyclic peptide with affinity for the ανβ3 integrin in human melanoma cells. Eur. J. Cancer 2000, 36, 410–422. [Google Scholar] [CrossRef]
- Guo, Y.; Lu, X.; Weng, J.; Leng, Y. Density functional theory study of the interaction of arginine-glycine-aspartic acid with graphene, defective graphene, and graphene oxide. J. Phys. Chem. C 2013, 117, 5708–5717. [Google Scholar] [CrossRef]
- Jiang, M.; Qi, L.; Liu, P.; Wang, Z.; Duan, Z.; Wang, Y.; Liu, Z.; Chen, P. Selective enrichment and desalting of hydrophilic peptides using graphene oxide. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1027, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Baweja, L.; Balamurugan, K.; Subramanian, V.; Dhawan, A. Effect of graphene oxide on the conformational transitions of amyloid beta peptide: A molecular dynamics simulation study. J. Mol. Graph. Model. 2015, 61, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Geim, A.K.; Novoselov, K.S. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.Y.; Chien, Y.; Chuang, J.H.; Chang, C.C.; Yang, Y.P.; Lai, Y.H.; Lo, W.L.; Chien, K.H.; Huo, T.I.; Wang, C.Y. Development of a graphene oxide-incorporated polydimethylsiloxane membrane with hexagonal micropillars. Int. J. Mol. Sci. 2018, 19, 2517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Deng, Y.S.; Tian, H.B.; Yan, H.; Cui, H.L.; Wang, D.Q. Noise analysis of monolayer graphene nanopores. Int. J. Mol. Sci. 2018, 19, 2639. [Google Scholar] [CrossRef] [PubMed]
- Imani, R.; Emami, S.H.; Faghihi, S. Synthesis and characterization of an octaarginine functionalized graphene oxide nanocarrier for gene delivery applications. Phys. Chem. Chem. Phys. 2015, 17, 6328–6339. [Google Scholar] [CrossRef] [PubMed]
- Kizil, S.; Bulbul Sonmez, H. Oil loving hydrophobic gels made from glycerol propoxylate: Efficient and reusable sorbents for oil spill clean-up. J. Environ. Manag. 2017, 196, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Pan, Y.; Ping, Y.; Sahoo, N.G.; Wu, T.; Li, L.; Li, J.; Gan, L.H. Chitosan-functionalized graphene oxide as a nanocarrier for drug and gene delivery. Small 2011, 7, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Shin, Y.C.; Lee, J.J.; Bae, E.B.; Jeon, Y.C.; Jeong, C.M.; Yun, M.J.; Lee, S.H.; Han, D.W.; Huh, J.B. The effect of reduced graphene oxide-coated biphasic calcium phosphate bone graft materialon osteogenesis. Int. J. Mol. Sci. 2018, 18, 1725. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, N.; Matchett, G.; Solaroglu, I.; Tsubokawa, T.; Ohkuma, H.; Zhang, J. Inhibition of integrin αvβ3 reduces blood-brain barrier breakdown in focal ischemia in rats. J. Neurosci. Res. 2006, 84, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, N.; Matchett, G.; Yatsushige, H.; Calvert, J.W.; Ohkuma, H.; Zhang, J. Inhibition of integrin αvβ3 ameliorates focal cerebral ischemic damage in the rat middle cerebral artery occlusion model. Stroke 2006, 37, 1902–1909. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, J.; Xie, K.; Wang, W.; Fang, Y. Aerobic oxidation of cyclohexane catalyzed by grapheneoxide:Effects of surface structure and functionalization. Mol. Catal. 2017, 431, 1–8. [Google Scholar] [CrossRef]
- Jafarizad, A.; Taghizadehgh-Alehjougi, A.; Eskandani, M.; Hatamzadeh, M.; Abbasian, M.; Mohammad-Rezaei, R.; Mohammadzadeh, M.; Toğar, B.; Jaymand, M. PEGylatedgraphene oxide/Fe3O4 nanocomposite: Synthesis, characterization, and evaluation of its performance as de novo drug delivery nanosystem. Biomed. Mater. Eng. 2018, 29, 177–190. [Google Scholar] [PubMed]
- Chen, J.; Zhang, X.; Cai, H.; Chen, Z.; Wang, T.; Jia, L.; Wang, J.; Wan, Q.; Pei, X. Osteogenic activity and antibacterial effect of zinc oxide/carboxylatedgraphene oxide nanocomposites: Preparation and in vitro evaluation. Colloids Surf. B 2016, 147, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Lelle, M.; Freidel, C.; Kaloyanova, S.; Tabujew, I.; Schramm, A.; Musheev, M.; Niehrs, C.; Müllen, K.; Peneva, K. Overcoming drug resistance by cell-penetrating peptide-mediated delivery of a doxorubicin dimer with high DNA-binding affinity. Eur. J. Med. Chem. 2017, 130, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Oda, H.; Inoue, Y.; Ishihara, K. Movement of a quantum dot covered with cytocompatible and ph responsible phospholipid polymer chains under a cellular environment. Biomacromolecules 2016, 17, 3986–3994. [Google Scholar] [CrossRef] [PubMed]
- Imani, R.; Shao, W.; Taherkhani, S.; Emami, S.H.; Prakash, S.; Faghihi, S. Dual-functionalized graphene oxide for enhanced siRNA delivery to breast cancer cells. Colloids Surf. B 2016, 147, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Ni, G.; Wang, Y.; Wu, X.; Wang, X.; Chen, S.; Liu, X. Graphene oxide absorbed anti-IL10R antibodies enhance LPS induced immune responses in vitro and in vivo. Immunol. Lett. 2012, 148, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, Z.; Deng, R.; Li, C.; Fu, S.; Chen, G.; Zhang, X.; Ke, F.; Ke, S.; Yu, X.; et al. Aptamer-mediated gene therapy enhanced antitumor activity against human hepatocellular carcinoma in vitro and in vivo. J. Control. Release 2017, 258, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Huang, W.; Jin, M.; Wang, Q.; Fan, B.; Kang, L.; Gao, Z. Chitosan-based nanoparticles for survivin targeted siRNA delivery in breast tumor therapy and preventing its metastasis. Int. J. Nanomed. 2016, 11, 4931–4945. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.W.; Kim, H.Y.; Li, F.; Park, J.Y.; Kim, D.; Park, J.H.; Han, H.S.; Byun, J.W.; Lee, Y.S.; Jeong, J.M.; et al. In vivo visualization of endogenous miR-21 using hyaluronic acidcoated graphene oxide for targeted cancer therapy. Biomaterials 2017, 121, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Schumann, C.; Chan, S.; Millar, J.A.; Bortnyak, Y.; Carey, K.; Fedchyk, A.; Wong, L.; Korzun, T.; Moses, A.S.; Lorenz, A.; et al. Intraperitonealnanotherapy for metastatic ovarian cancer based on siRNA-mediated suppression of DJ-1 protein combined with a low dose of cisplatin. Nanomedicine 2018, 14, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Sun, Y.; Zheng, S.L.; Qin, Y.; Mcclements, D.J.; Hu, J.N.; Deng, Z.Y. Antitumor and immunomodulatory effects of ginsenoside Rh2 and its octyl ester derivative in H22 tumor-bearing mice. J. Funct. Foods 2017, 32, 382–390. [Google Scholar] [CrossRef]
- Sohn, E.J.; Jung, D.B.; Lee, H.; Han, I.; Lee, J.; Lee, H.; Kim, S.H. CNOT2 promotes proliferation and angiogenesis via VEGF signaling in MDA-MB-231 breast cancer cells. Cancer Lett. 2018, 412, 88–98. [Google Scholar] [CrossRef] [PubMed]





















© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Ge, X.; Cui, C.; Zhang, Y.; Wang, Y.; Wang, X.; Sun, Q. Preparation and Characterization of Functionalized Graphene Oxide Carrier for siRNA Delivery. Int. J. Mol. Sci. 2018, 19, 3202. https://doi.org/10.3390/ijms19103202
Li J, Ge X, Cui C, Zhang Y, Wang Y, Wang X, Sun Q. Preparation and Characterization of Functionalized Graphene Oxide Carrier for siRNA Delivery. International Journal of Molecular Sciences. 2018; 19(10):3202. https://doi.org/10.3390/ijms19103202
Chicago/Turabian StyleLi, Jing, Xu Ge, Chunying Cui, Yifan Zhang, Yifan Wang, Xiaoli Wang, and Qi Sun. 2018. "Preparation and Characterization of Functionalized Graphene Oxide Carrier for siRNA Delivery" International Journal of Molecular Sciences 19, no. 10: 3202. https://doi.org/10.3390/ijms19103202
APA StyleLi, J., Ge, X., Cui, C., Zhang, Y., Wang, Y., Wang, X., & Sun, Q. (2018). Preparation and Characterization of Functionalized Graphene Oxide Carrier for siRNA Delivery. International Journal of Molecular Sciences, 19(10), 3202. https://doi.org/10.3390/ijms19103202