Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP
Abstract
:1. Introduction
2. Results
2.1. NopP Effects on Soybean Germplasm
2.2. Phenotypic Analysis
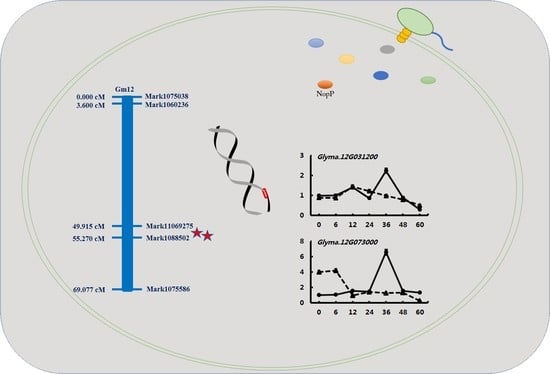
2.3. Mapping of Conditional QTL for Nodulation-Related Traits
2.4. Validation of Candidate Genes by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids and Primers
4.2. Generation of the HH103ΩNopP Mutant
4.3. Nodulation Tests
Plant Materials and Nodulation-Related Traits
4.4. The Conditional QTL Mapping of Nodulation-Related Traits
4.5. Delimitation of QTL Regions and Identification of Candidate Genes
4.6. RNA Isolation and qRT-PCR Analyses of NopP Candidate Genes
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xin, D.W.; Qi, Z.M.; Jiang, H.W.; Hu, Z.B.; Zhu, R.S.; Hu, J.H.; Han, H.Y.; Hu, G.H.; Liu, C.Y.; Chen, Q.S. QTL Location and Epistatic Effect Analysis of 100-Seed Weight Using Wild Soybean (Glycine soja Sieb. & Zucc.) Chromosome Segment Substitution Lines. PLoS ONE 2016, 11, e0149380. [Google Scholar] [CrossRef]
- Zimmer, S.; Messmer, M.; Haase, T.; Piepho, H.P.; Mindermann, A.; Schulz, H.; Habekuß, A.; Ordon, F.; Wilbois, P.H.; Heß, J. Effects of soybean variety and Bradyrhizobium strains on yield, protein content and biological nitrogen fixation under cool growing conditions in Germany. Eur. J. Agron. 2016, 72, 38–46. [Google Scholar] [CrossRef]
- Riely, B.K.; Larrainzar, E.; Haney, C.H.; Mun, J.H.; Gil-Quintana, E.; González, E.M.; Yu, H.J.; Tricoli, D.; Ehrhardt, D.W.; Cook, D.R. Development of tools for the biochemical characterization of the symbiotic receptor-like kinase DMI2. Mol. Plant Microbe Interact. 2013, 26, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Riches, D.; Porter, I.J.; Oliver, D.P.; Bramley, R.G.V.; Rawnsley, B.; Edwards, J.; White, R.E. Soil biological properties as indicators of soil quality in A ustralian viticulture. Aust. J. Grape Wine Res. 2003, 19, 311–323. [Google Scholar] [CrossRef]
- Buendía-Clavería, A.M.; Moussaid, A.; Ollero, F.J.; Vinardell, J.M.; Torres, A.; Moreno, J.; Gil-Serrano, A.M.; Rodríguez-Carvajal, M.A.; Tejero-Mateo, P.; Peart, J.L. A purL mutant of Sinorhizobium fredii HH103 is symbiotically defective and altered in its lipopolysaccharide. Microbiology 2003, 149, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Yan, G.H.; Li, J.L. Numerical taxonomy study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Evol. Microbiol. 1988, 38, 392–397. [Google Scholar] [CrossRef]
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial Molecular Signals in the Sinorhizobium fredii-Soybean Symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 nodulation outer protein NopI is a determinant for efficient nodulation of soybean and cowpea plants. Appl. Environ. Microbiol. 2016, 83, e02770-16. [Google Scholar] [CrossRef] [PubMed]
- Margaret, I.; Becker, A.; Blom, J.; Bonilla, I.; Goesmann, A.; Göttfert, M.; Lloret, J.; Mittard-Runte, V.; Rückert, C.; Ruiz-Sainz, J.E.; et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011, 155, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Weidner, S.; Becker, A.; Bonilla, I.; Jaenicke, S.; Lloret, J.; Margaret, I.; Pühler, A.; Ruiz-Sainz, J.E.; Schneiker-Bekel, S.; Szczepanowski, R.; et al. Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J. Biotechnol. 2012, 194, 1617–1618. [Google Scholar] [CrossRef] [PubMed]
- Vinardell, J.M.; Acostajurado, S.; Zehner, S.; Göttfert, M.; Becker, A.; Baena, I.; Blom, J.; Goesmann, A.; Jaenicke, S.; Krol, E.; et al. The Sinorhizobium fredii HH103 genome: A comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol. Plant Microbe Interact. 2015, 28, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Acosta-Jurado, S.; Navarro-Gómez, P.; Ollero, F.J.; Ruiz-Sainz, J.E.; López-Baena, F.J.; Vinardell, J.M. A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 2016, 19, 31592. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, L.Y.; Liu, W.; Tian, Y.; Xiong, J.S.; Wang, Y.H.; Li, R.J.; Li, H.M.; Wen, J.Q.; Mysore, K.S.; et al. Role of the Nod factor hydrolase MtNFH1 in regulating Nod factor levels during rhizobial infection and in mature nodules of Medicago truncatula. Plant Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Gourion, B.; Berrabah, F.; Ratet, P.; Stacey, G. Rhizobium–legume symbioses: The crucial role of plant immunity. Trends Plant Sci. 2015, 20, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Gully, D.; Gargani, D.; Bonaldi, K.; Grangeteau, C.; Chaintreuil, C.; Fardoux, J.; Nguyen, P.; Marchetti, R.; Nouwen, N.; Molinaro, A. A peptidoglycan-remodeling enzyme is critical for bacteroid differentiation in Bradyrhizobium spp. during legume symbiosis. Mol. Plant Microbe Interact. 2016, 29, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yang, S.; Tang, F.; Zhu, H. Symbiosis specificity in the legume–rhizobial mutualism. Cell Microbiol. 2012, 14, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.W.; Thygesen, M.B.; Snadal, N.; Asmussen, M.H. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Halane, M.K.; Gassmann, W.; Stacey, G. The role of plant innate immunity in the legume-rhizobium symbiosis. Ann. Rev. Plant Biol. 2017, 68, 535–561. [Google Scholar] [CrossRef] [PubMed]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within symbiosis: Evolving nitrogen-fixing legume symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Deakin, W.J.; Viprey, V.; Kopciñska, J.; Golinowski, W.; Krishnan, H.B.; Perret, X.; Broughton, W.J. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 2003, 16, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Tampakaki, A.P. Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front Plant Sci. 2014, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, C.; Krishnan, H.B. Nodulation outer proteins: Double-edged swords of symbiotic rhizobia. Biochem. J. 2015, 470, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, H.B. NolX of Sinorhizobium fredii USDA257, a type III-secreted protein involved in host range determination, is localized in the infection threads of cowpea (Vigna unguiculata [L.] Walp) and soybean (Glycine max [L.] Merr.) nodules. J. Bacteriol. 2002, 184, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Bartsev, A.V.; Boukli, N.M.; Deakin, W.J.; Staehelin, C.; Broughton, W.J. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 2003, 554, 271–274. [Google Scholar] [CrossRef]
- Ausmees, N.; Kobayashi, H.; Deakin, W.J.; Marie, C.; Krishnan, H.B.; Broughton, W.J.; Perret, X. Characterization of NopP, a type III secreted effector of Rhizobium sp. strain NGR234. J. Bacteriol. 2004, 186, 4774–4780. [Google Scholar] [CrossRef] [PubMed]
- Lorio, J.C.; Kim, W.S.; Krishnan, H.B. NopB, a soybean cultivar-specificity protein from Sinorhizobium fredii USDA257, is a type III secreted protein. Mol. Plant Microbe Interact. 2004, 17, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Deakin, W.J.; Marie, C.; Saad, M.M.; Krishnan, H.B.; Broughton, W.J. NopA is associated with cell surface appendages produced by the type III secretion system of Rhizobium sp. strain NGR234. Mol. Plant Microbe Interact. 2005, 18, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Skorpil, P.; Saad, M.M.; Boukli, N.M.; Kobayashi, H.; Ares-Orpel, F.; Broughton, W.J.; Deakin, W.J. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 2005, 57, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.A.; Lopez-Baena, F.J.; Ollero, F.J.; Vinardell, J.M.; Espuny, M.D.; Bellogín, R.A.; Thomas, J.R.; Sumpton, D.; Ault, J.; Thomas-Oates, J. NopM and NopD are rhizobial nodulation outer proteins: Identification using LC-MALDI and LC-ESI with a monolithic capillary column. J. Proteome Res. 2007, 6, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.J.; Zeng, Y.; Xie, Z.P.; Staehelin, C. Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of Rhizobium sp. strain NGR234. J. Bacteriol. 2008, 190, 5101–5110. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.M.; Staehelin, C.; Broughton, W.J.; Deakin, W.J. Protein-protein interactions within type III secretion system-dependent pili of Rhizobium sp. strain NGR234. J. Bacteriol. 2008, 190, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Kambara, K.; Ardissone, S.; Kobayashi, H.; Saad, M.M.; Schumpp, O.; Broughton, W.J.; Deakin, W.J. Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 2009, 71, 92–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Chen, X.J.; Lu, H.B.; Xie, Z.P.; Staehelin, C. Functional analysis of the type 3 effector NopL from Rhizobium sp. NGR234: Symbiotic effects, phosphorylation and interference with MAPK signaling. J. Biol. Chem. 2011, jbc-M111, 265942. [Google Scholar] [CrossRef]
- Xin, D.W.; Liao, S.; Xie, Z.P.; Hann, D.R.; Steinle, L.; Boller, T.; Staehelin, C. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 2012, 8, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Kimbrel, J.A.; Thomas, W.J.; Jiang, Y.; Creason, A.L.; Thireault, C.A.; Sachs, J.L.; Chang, J.H. Mutualistic co-evolution of type III effector genes in Sinorhizobium fredii and Bradyrhizobium japonicum. PLoS Pathog. 2013, 9, e1003204. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.Y.; Xiang, Q.W.; Wagner, C.; Zhang, D.; Xie, Z.P.; Staehelin, C. The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 2016, 67, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC is a Rhizobium-specific type 3 secretion system effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, e0142866. [Google Scholar] [CrossRef] [PubMed]
- Schechter, L.M.; Guenther, J.; Olcay, E.A.; Jang, S.; Krishnan, H.B. Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl. Environ. Microbiol. 2010, 76, 3758–3761. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Monreal, J.A.; Pérez-Montaño, F.; Guasch-Vidal, B.; Bellogín, R.A.; Vinardell, J.M.; Ollero, F.J. The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams soybean. Mol. Plant Microbe Interact. 2009, 22, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Hajimorad, M.R.; Hill, J.H. Rsv1-mediated resistance against Soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant Microbe Interact. 2001, 14, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Penninckx, I.A.; Cammue, B.P.; Broekaert, W.F. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001, 13, 63–68. [Google Scholar] [CrossRef]
- Jones, D.A.; Takemoto, D. Plant innate immunity–direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 2004, 16, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Saeki, Y.; Haga, M.; Harada, K.; Kouchi, H.; Umehara, Y. Rj (rj) genes involved in nitrogen-fixing root nodule formation in soybean. Breeding Sci. 2012, 61, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, B.E.; Vest, G. Nodulation interactions between soybean genotypes and serogroups of Rhizobium japonicum. Crop Sci. 1968, 8, 680–682. [Google Scholar] [CrossRef]
- Tsukui, T.; Eda, S.; Kaneko, T.; Sato, S.; Okazaki, S.; Kakizaki-Chiba, K.; Itakura, M.; Mitsui, H.; Yamashita, A.; Terasawa, K.; et al. The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 2013, 79, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, M.; Takahashi, S.; Umehara, Y.; Iwano, H.; Tsurumaru, H.; Odake, H.; Suzuki, Y.; Kondo, H.; Konno, Y.; Yamakawa, T.; et al. Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity. Nat. Commun. 2018, 9, 3139. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Ray, J.D.; Cregan, P.B.; King, C.A.; Davies, M.K.; Purcell, L.C. Genetics and mapping of quantitative traits for nodule number, weight, and size in soybean (Glycine max L.[Merr.]). Euphytica 2014, 195, 419–434. [Google Scholar] [CrossRef]
- Santos, M.A.; Geraldi, I.O.; Garcia, A.A.F.; Bortolatto, N.; Schiavon, A.; Hungria, M. Mapping of QTLs associated with biological nitrogen fixation traits in soybean. Hereditas 2013, 150, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caetano-Anolles, G.; Gresshoff, P.M. Plant genetic control of nodulation. Ann. Rev. Microbiol. 1991, 45, 345–382. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, B.E. Inheritance of a Strain-Specific Ineffective Nodulation in Soybeans. Crop Sci. 1966, 6, 427–428. [Google Scholar] [CrossRef]
- Vest, G. Rj3 A Gene Conditioning Ineffective Nodulation in Soybean. Crop Sci. 1970, 10, 34–35. [Google Scholar] [CrossRef]
- Vest, G.; Caldwell, B.E. Rj4—A Gene Conditioning Ineffective Nodulation in Soybean. Crop Sci. 1972, 12, 692–693. [Google Scholar] [CrossRef]
- Vuong, T.D.; Nickell, C.D.; Harper, J.E. Genetic and allelism analyses of hypernodulation soybean mutants from two genetic backgrounds. Crop Sci. 1996, 36, 1153–1158. [Google Scholar] [CrossRef]
- Yang, S.M.; Tang, F.; Gao, M.Q.; Krishnan, H.B.; Zhu, H.Y. R gene-controlled host specificity in the legume–rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 2010, 201011957. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Yang, S.M.; Liu, J.G.; Zhu, H.Y. Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 2016, 170, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 201302360. [Google Scholar] [CrossRef] [PubMed]
- Faruque, O.M.; Miwa, H.; Yasuda, M.; Fujii, Y.; Kaneko, T.; Sato, S.; Okazaki, S. Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 2015, 81, 6710–6717. [Google Scholar] [CrossRef] [PubMed]
- Viprey, V.; Del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.J.; Liu, X.Y.; Chen, L.; Fu, Y.; Li, C.Y.; Qi, Z.M.; Zou, J.N.; Zhu, R.S.; Li, S.P.; Wei, W.; et al. Mining for genes encoding proteins associated with NopL of Sinorhizobium fredii HH103 using quantitative trait loci in soybean (Glycine max Merr.) recombinant inbred lines. Plant Soil 2018, 431, 245–255. [Google Scholar] [CrossRef]
- Edens, L.; Heslinga, L.; Klok, R.; Ledeboer, M.A.; Maat, J.; Toonen, M.Y.; Visser, C.; Theo Verrips, C. Cloning of cDNA encoding the sweet-tasting plant protein thaumatin and its expression in Escherichia coli. Gene 1982, 18, 1–12. [Google Scholar] [CrossRef]
- Wang, X.J.; Tang, C.L.; Deng, L.; Cai, G.L.; Liu, X.Y.; Liu, B.; Han, Q.M.; Buchenauer, H.; Wei, G.R.; Han, D.Y.; et al. Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust fungus. Physiol. Plant. 2010, 139, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.C.; Bushnell, W.R.; Szabo, L.J.; Smith, A.G. Isolation and expression of a host response gene family encoding thaumatin-like proteins in incompatible oat-stem rust fungus interactions. Mol. Plant Microbe Interact. 1996, 9, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Reddy, A.S.N. Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Mol. Biol. 1997, 34, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Fukumoto, T.; Tatano, S.; Gomi, K.; Ohtani, K.; Tada, Y.; Akimitsu, K. Molecular cloning and characterization of a thaumatin-like protein-encoding cDNA from rough lemon. Physiol. Mol. Plant Pathol. 2009, 74, 3–10. [Google Scholar] [CrossRef]
- Okazaki, S.; Zehner, S.; Hempel, J.; Lang, K.; Göttfert, K. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009, 295, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Neupane, A.; Nepal, M.P.; Benson, B.V.; MacArthur, K.J.; Piya, S. Evolutionary history of mitogen-activated protein kinase (MAPK) genes in Lotus, Medicago, and Phaseolus. Plant Signal. Behav. 2013, 8, e27189. [Google Scholar] [CrossRef] [PubMed]
- Grimsrud, P.A.; den Os, D.; Wenger, C.D.; Swaney, D.L.; Schwartz, D.; Sussman, M.R.; Ané, J.M.; Coon, J.J. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 2010, 152, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Declerck, M.; Sorin, C.; Hartmann, C.; Crespi, M.; Lelandais-Brière, C. MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 2011, 77, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Schoenbeck, M.A.; Samac, D.A.; Fedorova, M.; Gregerson, R.G.; Gantt, J.S.; Vance, C.P. The alfalfa (Medicago sativa) TDY1 gene encodes a mitogen-activated protein kinase homolog. Mol. Plant Microbe Interact. 1999, 12, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pascual, M.; Lucas, M.M.; de Felipe, M.R.; Boscá, L.; Hirt, H.; Golvano, M.P. Involvement of mitogen-activated protein kinases in the symbiosis Bradyrhizobium–Lupinus. J. Exp. Bot. 2006, 57, 2735–2742. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kim, J.; Im, J.H.; Kim, H.B.; Oh, C.J.; An, C.S. Mitogen-activated protein kinase is involved in the symbiotic interaction between Bradyrhizobium japonicum USDA110 and soybean. J. Plant Biol. 2008, 51, 291–296. [Google Scholar] [CrossRef]
- Trosky, J.E.; Mukherjee, S.; Burdette, D.L.; Roberts, M.; McCarter, L.; Siegel, R.M.; Orth, K. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J. Biol. Chem. 2004, 279, 51953–51957. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Ann. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Kachroo, P.; Tsui, F.; Sharma, S.B.; Shah, J.; Klessig, D.F. Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 2001, 26, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Miwa, H.; Masuda, S.; Takebayashi, Y.; Sakakibara, H.; Okazaki, S. Effector-triggered immunity determines host genotype-specific incompatibility in legume–Rhizobium symbiosis. Plant Cell Physiol. 2016, 57, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Quandt, J.; Hynes, M.F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 1993, 127, 15–21. [Google Scholar] [CrossRef]
- Figurski, D.; Meyer, R.J.; Helinski, D.R. Suppression of ColE1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J. Mol. Biol. 1979, 133, 295–318. [Google Scholar] [CrossRef]
- Jiang, H.W.; Li, Y.Y.; Qin, H.T.; Li, Y.L.; Qi, H.D.; Li, C.D.; Wang, N.N.; Li, R.C.; Zhao, Y.Y.; Huang, S.Y. Identification of Major QTLs Associated With First Pod Height and Candidate Gene Mining in Soybean. Front Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Brensha, W.; Kantartzi, S.K.; Meksem, K.; Grier IV, R.L.; Barakat, A.; Lightfoot, D.A.; Kassem, M.A. Genetic analysis of root and shoot traits in the ‘Essex’ by ‘Forrest’ recombinant inbred line (RIL) population of soybean [Glycine max (L.) Merr.]. Plant Genet. Genom. Biotechnol. 2012, 1, 1–9. [Google Scholar] [CrossRef]
- Qi, Z.M.; Huang, L.; Zhu, R.S.; Xin, D.W.; Liu, C.Y.; Han, X.; Jiang, H.W.; Chen, Q.S. A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS ONE 2014, 9, e104871. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [PubMed]





| RILs (n = 150) | Parents (Average) | |||||
|---|---|---|---|---|---|---|
| Traits | Average | Standard Deviation | Coefficient of Variation | Charleston | Dongnong594 | |
| HH103 RifR | Nodule number | 23.9 | 15.1 | 76.38 | 14.0 ± 3.5 ** | 26.0 ± 6.6 |
| Nodule dry-weight (mg) | 18.4 * | 17.2 | 93.25 | 13.0 ± 2.6 ** | 22.5 ± 1.8 | |
| HH103 RifRΩNopP | Nodule number | 11.5 ** | 13.822 | 58.07 | 13.0 ± 3.0 ** | 27.0 ± 3.0 |
| Nodule dry-weight (mg) | 29.4 | 0.0236 | 80.34 | 10.0 ± 2.6 ** | 41.5 ± 5.6 | |
| Trait | LG/QTL | Chrom. | Position (cM) | LOD a | R2 (%) b | ADD c |
|---|---|---|---|---|---|---|
| Nodule number | QN/NN01 | 03 | 55.5 | 3.8 | 8.21 | −2.46 |
| QB2/NN02 | 14 | 74.0 | 4.6 | 9.04 | −2.95 | |
| QD2/NN03 | 17 | 133.2 | 5.0 | 8.19 | 2.61 | |
| Nodule dry weight | QN/NDW01 | 03 | 74.0 | 5.0 | 2.54 | −0.001 |
| QH/NDW02 | 12 | 51.8 | 4.0 | 9.78 | 0.005 | |
| QH/NDW03 | 12 | 55.5 | 5.4 | 3.75 | 0.002 | |
| QJ/NDW04 | 16 | 88.8 | 4.5 | 5.26 | −0.002 |
| No. | Gene | Function |
|---|---|---|
| 1 | Glyma.12G028300 | Cell elongation protein/DWARF1/diminuto (DIM) |
| 2 | Glyma.12G030000 | Leucine-rich repeat protein kinase family protein |
| 3 | Glyma.12G030300 | Disease resistance-responsive (dirigent-like protein) family protein |
| 4 | Glyma.12G030900 | Like auxin resistant 2 |
| 5 | Glyma.12G031000 | Pathogenesis-related thaumatin superfamily protein |
| 6 | Glyma.12G031200 | Pathogenesis-related thaumatin superfamily protein |
| 7 | Glyma.12G032400 | Calcium-dependent lipid-binding (CaLB domain) family protein |
| 8 | Glyma.12G036700 | NAD(P)-linked oxidoreductase superfamily protein |
| 9 | Glyma.12G036900 | NAD(P)-binding rossmann-fold superfamily protein |
| 10 | Glyma.12G040000 | Leucine-rich receptor-like protein kinase family protein |
| 11 | Glyma.12G043600 | Protein kinase superfamily protein |
| 12 | Glyma.12G052400 | Protein kinase family protein |
| 13 | Glyma.12G055500 | Leucine-rich repeat (LRR) family protein |
| 14 | Glyma.12G055600 | Leucine-rich repeat (LRR) family protein |
| 15 | Glyma.12G073000 | Mitogen-activated protein kinase 3 |
| 16 | Glyma.12G076200 | Auxin response factor 10 |
| 17 | Glyma.12G079300 | SAUR-like auxin-responsive protein family |
| No. | Gene | Function |
|---|---|---|
| 1 | Glyma.12G028300 | Cell elongation protein/DWARF1/Diminuto (DIM) |
| 2 | Glyma.12G030000 | Leucine-rich repeat protein kinase family protein |
| 3 | Glyma.12G031200 | Pathogenesis-related thaumatin superfamily protein |
| 4 | Glyma.12G036900 | NAD(P)-binding rossmann-fold superfamily protein |
| 5 | Glyma.12G052400 | Protein kinase family protein |
| 6 | Glyma.12G055500 | Leucine-rich repeat (LRR) family protein |
| 7 | Glyma.12G073000 | Mitogen-activated protein kinase 3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Wang, J.; Liu, C.; Ma, C.; Li, C.; Zhang, Y.; Qi, Z.; Zhu, R.; Shi, Y.; Zou, J.; et al. Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. Int. J. Mol. Sci. 2018, 19, 3438. https://doi.org/10.3390/ijms19113438
Wang J, Wang J, Liu C, Ma C, Li C, Zhang Y, Qi Z, Zhu R, Shi Y, Zou J, et al. Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. International Journal of Molecular Sciences. 2018; 19(11):3438. https://doi.org/10.3390/ijms19113438
Chicago/Turabian StyleWang, Jinhui, Jieqi Wang, Chunyan Liu, Chao Ma, Changyu Li, Yongqian Zhang, Zhaoming Qi, Rongsheng Zhu, Yan Shi, Jianan Zou, and et al. 2018. "Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP" International Journal of Molecular Sciences 19, no. 11: 3438. https://doi.org/10.3390/ijms19113438
APA StyleWang, J., Wang, J., Liu, C., Ma, C., Li, C., Zhang, Y., Qi, Z., Zhu, R., Shi, Y., Zou, J., Li, Q., Zhu, J., Wen, Y., Sun, Z., Liu, H., Jiang, H., Yin, Z., Hu, Z., Chen, Q., ... Xin, D. (2018). Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. International Journal of Molecular Sciences, 19(11), 3438. https://doi.org/10.3390/ijms19113438