Exercise Training-Induced Changes in MicroRNAs: Beneficial Regulatory Effects in Hypertension, Type 2 Diabetes, and Obesity
Abstract
:1. Introduction
1.1. MicroRNAs
1.2. MiRNAs as Mediators and Biomarkers of Cardiovascular Diseases
2. MiRNAs and Systemic Arterial Hypertension (SAH)
2.1. MiRNAs and Endothelial Dysfunction in SAH
2.2. MiRNAs and Arterial Remodeling in SAH
2.3. MiRNAs and Renin-Angiotensin-Aldosterone System (RAAS) in SAH
2.4. MiRNAs and Autonomic Nervous System in SAH
2.5. Circulating miRNAs in SAH
2.6. Single Nucleotide Polymorphisms and miRNAs in SAH
3. MiRNAs and Type 2 Diabetes Mellitus
3.1. MiRNAs, Insulin Synthesis, and Secretion
3.2. MiRNAs and Insulin Resistance
3.3. MiRNAs and Lipid Metabolism
3.4. Circulating miRNAs in T2D
3.5. Single Nucleotide Polymorphisms and miRNAs in T2D
4. MiRNAs and Obesity
4.1. MiRNAs Involved with Adipocyte Differentiation and Proliferation
4.2. Other Circulating miRNAs in Obesity
4.3. Single Nucleotide Polymorphisms and miRNAs in Obesity
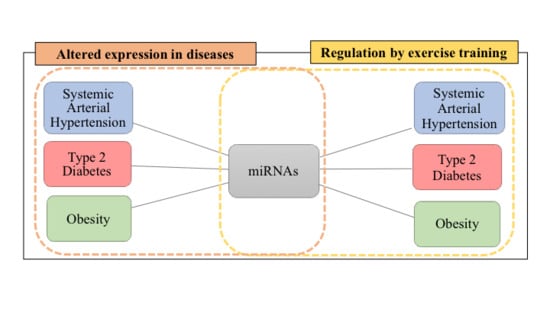
5. Overlapping miRNAs in SAH, T2D, and Obesity
6. Exercise and Cardiovascular Protection
6.1. Altered miRNA Expression Induced by Exercise Training
6.2. Overlaps Between the miRNA Profile in SAH, T2D, Obesity, and Exercise
6.3. SAH vs. Exercises: miRNAs Differentially Expressed
6.4. T2D versus Exercises: miRNAs Differentially Expressed
6.5. Obesity vs. Exercises: miRNAs Differentially Expressed
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; Van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Muljo, S.A.; Kanellopoulou, C.; Aravind, L. MicroRNA targeting in mammalian genomes: Genes and mechanisms. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Kawai-Kowase, K.; Owens, G.K. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2007, 292, C59–C69. [Google Scholar] [CrossRef] [PubMed]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Krüger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Investig. 2009, 119, 2634–2647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, W.; Xiao, D.; Ming, G.; Yin, J.; Zhou, H.; Liu, Z. Type 2 diabetes mellitus-related genetic polymorphisms in microRNAs and microRNA target sites. J. Diabetes 2014, 6, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Marioni, J.C.; Pai, A.A.; Degner, J.F.; Engelhardt, B.E.; Nkadori, E.; Veyrieras, J.B.; Stephens, M.; Gilad, Y.; Pritchard, J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 2010, 464, 768–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Gong, Y.; Sun, A.; Zhang, Y.; Zhang, C.; Zhang, W.; Zhao, G.; Zou, Y.; Ge, J. The human MTHFR rs4846049 polymorphism increases coronary heart disease risk through modifying miRNA binding. Nutr. Metab. Cardiovasc. Dis. 2013, 23, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Nyayanit, D.; Gadgil, C.J. Mathematical modeling of combinatorial regulation suggests that apparent positive regulation of targets by miRNA could be an artifact resulting from competition for mRNA. RNA 2015, 21, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, C. MicroRNA-21 in cardiovascular disease. J. Cardiovasc. Transl. Res. 2010, 3, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Sato-Kuwabara, Y.; Melo, S.A.; Soares, F.A.; Calin, G.A. The fusion of two worlds: Non-coding RNAs and extracellular vesicles--diagnostic and therapeutic implications (Review). Int. J. Oncol. 2015, 46, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Yip, K.W.; Spence, T.; Liu, F.F. MicroRNAs in extracellular vesicles: Potential cancer biomarkers. J. Hum. Genet. 2017, 62, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 23715. [Google Scholar] [CrossRef] [PubMed]
- Go, A.S.; Mozaffarian, D.; Roger, V.L.; Benjamin, E.J.; Berry, J.D.; Blaha, M.J.; Dai, S.; Ford, E.S.; Fox, C.S.; Franco, S.; et al. Executive summary: Heart disease and stroke statistics--2014 update: A report from the American Heart Association. Circulation 2014, 129, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.S.; Brownlee, M. Molecular and cellular mechanisms of cardiovascular disorders in diabetes. Circ. Res. 2016, 118, 1808–1829. [Google Scholar] [CrossRef] [PubMed]
- IDF Diabetes Atlas. Available online: https://www.idf.org/e-library/epidemiology-research/diabetes-atlas/13-diabetes-atlas-seventh-edition.html (accessed on 29 June 2018).
- Mandavia, C.H.; Aroor, A.R.; Demarco, V.G.; Sowers, J.R. Molecular and metabolic mechanisms of cardiac dysfunction in diabetes. Life Sci. 2013, 92, 601–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogden, C.L.; Carroll, M.D.; Kit, B.K.; Flegal, K.M. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA 2014, 311, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. What we talk about when we talk about fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Kusminski, C.M.; Bickel, P.E.; Scherer, P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 2016, 15, 639–660. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.; Mejhert, N.; Kulyté, A.; Balwierz, P.J.; Pachkov, M.; Cormont, M.; Lorente-Cebrián, S.; Ehrlund, A.; Laurencikiene, J.; Hedén, P.; et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 2012, 61, 1986–1993. [Google Scholar] [CrossRef] [PubMed]
- Tanti, J.F.; Ceppo, F.; Jager, J.; Berthou, F. Implication of inflammatory signaling pathways in obesity-induced insulin resistance. Front. Endocrinol. (Lausanne) 2013, 3, 181. [Google Scholar] [CrossRef] [PubMed]
- Neves, V.J.; Fernandes, T.; Roque, F.R.; Soci, U.P.; Melo, S.F.; de Oliveira, E.M. Exercise training in hypertension: Role of microRNAs. World J. Cardiol. 2014, 6, 713–727. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO A Global Brief on Hypertension; WHO Press: Geneva, Switzerland, 2013; p. 40. [Google Scholar]
- Malachias, M.; Plavnik, F.L.; Machado, C.A.; Malta, D.; Scala, L.C.N.; Fuchs, S. 7th Brazilian Guideline of Arterial Hypertension: Chapter 1—Concept, Epidemiology and Primary Prevention. Arq. Bras. Cardiol. 2016, 107, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, J. The importance of drug combinations for effective control of hypertension. Clin. Exp. Hypertens. 1999, 21, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Lifton, R.P. Molecular genetics of human blood pressure variation. Science 1996, 272, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.T.; Boerwinkle, E. Genetics of blood pressure, hypertensive complications, and antihypertensive drug responses. Pharmacogenomics 2003, 4, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Kroll, M.H.; Schafer, A.I. Biochemical mechanisms of platelet activation. Blood 1989, 74, 1181–1195. [Google Scholar] [PubMed]
- Bassenge, E.; Huckstorf, C. Endothelium-dependent mechanisms of vascular regulation. Verh. Dtsch. Ges. Inn. Med. 1991, 97, 567–573. [Google Scholar] [PubMed]
- Li, Q.; Youn, J.Y.; Cai, H. Mechanisms and consequences of endothelial nitric oxide synthase dysfunction in hypertension. J. Hypertens. 2015, 33, 1128–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemecz, M.; Alexandru, N.; Tanko, G.; Georgescu, A. Role of microRNA in endothelial dysfunction and hypertension. Curr. Hypertens. Rep. 2016, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Manresa, N.; Mulero, J.; Losada, M.; Zafrilla, P. Influence of anti-VEGF about cardiovascular biomarkers in age related macular degeneration. J. Nutr Health Aging 2015, 19, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Tu, G.; Yang, G.; Li, G.; Yang, D.; Lang, L.; Xi, L.; Sun, K.; Chen, Y.; Shu, K.; et al. MiR-205/YAP1 in activated fibroblasts of breast tumor promotes VEGF-independent angiogenesis through STAT3 signaling. Theranostics 2017, 7, 3972–3988. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Diez, C.; Wilson, R.; Chatterjee, N.; Hudson, C.; Ruhl, R.; Hipfinger, C.; Helms, E.; Khan, O.F.; Anderson, D.G.; Anand, S. MicroRNA regulation of the MRN complex impacts DNA damage, cellular senescence, and angiogenic signaling. Cell Death Dis. 2018, 9, 632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Z.; Zhang, D.Y.; Zhu, J.; Zhang, T.; Wang, C. microRNA 126 inhibits the transition of endothelial progenitor cells to mesenchymal cells via the PIK3R2-PI3K/Akt signalling pathway. PLoS ONE 2013, 8, e83294. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: Potential markers of target-organ damage. J. Am. Soc. Hypertens. 2014, 8, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, Z.; Lv, Q.; Ye, W.; Wong, C.K.; Cai, G.; Gu, D.; Ji, Y.; Zhao, C.; Wang, J.; Yang, B.B.; et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE 2006, 1, e116. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jia, C.; Wang, P.; Xiong, M.; Cui, J.; Li, L.; Wang, W.; Wu, Q.; Chen, Y.; Zhang, T. MicroRNA-505 identified from patients with essential hypertension impairs endothelial cell migration and tube formation. Int. J. Cardiol. 2014, 177, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Palao, T.; Swärd, K.; Jongejan, A.; Moerland, P.D.; de Vos, J.; Van Weert, A.; Arribas, S.M.; Groma, G.; Van Bavel, E.; Bakker, E.N. Gene Expression and microRNA expression analysis in small arteries of spontaneously hypertensive rats. Evidence for er stress. PLoS ONE 2015, 10, e0137027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staszel, T.; Zapała, B.; Polus, A.; Sadakierska-Chudy, A.; Kieć-Wilk, B.; Stępień, E.; Wybrańska, I.; Chojnacka, M.; Dembińska-Kieć, A. Role of microRNAs in endothelial cell pathophysiology. Pol. Arch. Med. Wewn. 2011, 121, 361–366. [Google Scholar] [PubMed]
- Suárez, Y.; Wang, C.; Manes, T.D.; Pober, J.S. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: Feedback control of inflammation. J. Immunol. 2010, 184, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.A.; Carneiro, F.S.; Matsumoto, T.; Tostes, R.C. Bonus effects of anti-diabetic drugs: Possible beneficial effects on endothelial dysfunction, vascular inflammation and atherosclerosis. Basic Clin. Pharmacol. Toxicol. 2018, 123, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Huang, W.; Peng, J.; Zhu, T.T.; Sun, X.L.; Zhou, X.Y.; Yang, H.; Xiong, J.F.; He, H.Q.; Xu, Y.H.; et al. Irisin alleviates advanced glycation end products-induced inflammation and endothelial dysfunction via inhibiting ROS-NLRP3 inflammasome signaling. Inflammation 2018, 41, 260–275. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.X.; Zeng, D.Y.; Li, R.T.; Pang, R.P.; Yang, H.; Hu, Y.L.; Zhang, Q.; Jiang, Y.; Huang, L.Y.; Tang, Y.B.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Pankratz, F.; Bemtgen, X.; Zeiser, R.; Leonhardt, F.; Kreuzaler, S.; Hilgendorf, I.; Smolka, C.; Helbing, T.; Hoefer, I.; Esser, J.S.; et al. MicroRNA-155 exerts cell-specific antiangiogenic but proarteriogenic effects during adaptive neovascularization. Circulation 2015, 131, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Wang, X.; Wang, Y.; Tang, Z.; Cui, Q.; Xi, J.; Li, Y.S.; Chien, S.; Wang, N. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. USA 2010, 107, 3240–3244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Zhang, Y.C.; Chen, Y.; Xiang, Y.; Shen, C.X.; Li, Y.G. The role of miR-19b in the inhibition of endothelial cell apoptosis and its relationship with coronary artery disease. Sci. Rep. 2015, 5, 15132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, N.; Zhang, J.; Tong, Y. Hsa-let-7g miRNA targets caspase-3 and inhibits the apoptosis induced by ox-LDL in endothelial cells. Int. J. Mol. Sci. 2013, 14, 22708–22720. [Google Scholar] [CrossRef] [PubMed]
- White, K.; Dempsie, Y.; Caruso, P.; Wallace, E.; McDonald, R.A.; Stevens, H.; Hatley, M.E.; Van Rooij, E.; Morrell, N.W.; MacLean, M.R.; et al. Endothelial apoptosis in pulmonary hypertension is controlled by a microRNA/programmed cell death 4/caspase-3 axis. Hypertension 2014, 64, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liang, H.; Liu, H.; Li, D.; Chen, X.; Li, L.; Zhang, C.Y.; Zen, K. Platelet-secreted microRNA-223 promotes endothelial cell apoptosis induced by advanced glycation end products via targeting the insulin-like growth factor 1 receptor. J. Immunol. 2014, 192, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.A.; Semus, H.M.; Montgomery, R.L.; Stack, C.; Latimer, P.A.; Lewton, S.M.; Lynch, J.M.; Hullinger, T.G.; Seto, A.G.; Van Rooij, E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013, 15, 650–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.H.; Hu, C.P.; Chen, X.P.; Zhang, W.F.; Li, X.W.; Xiong, X.M.; Li, Y.J. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am. J. Hypertens. 2011, 24, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem. 2009, 284, 3728–3738. [Google Scholar] [CrossRef] [PubMed]
- Mandraffino, G.; Imbalzano, E.; Sardo, M.A.; D’Ascola, A.; Mamone, F.; Lo Gullo, A.; Alibrandi, A.; Loddo, S.; Mormina, E.; David, A.; et al. Circulating progenitor cells in hypertensive patients with different degrees of cardiovascular involvement. J. Hum. Hypertens. 2014, 28, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Carr, G.; Barrese, V.; Stott, J.B.; Povstyan, O.V.; Jepps, T.A.; Figueiredo, H.B.; Zheng, D.; Jamshidi, Y.; Greenwood, I.A. MicroRNA-153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension. Cardiovasc. Res. 2016, 112, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. Differential expression of vascular smooth muscle-modulating microRNAs in human peripheral blood mononuclear cells: Novel targets in essential hypertension. J. Hum. Hypertens. 2014, 28, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Elia, L.; Quintavalle, M.; Zhang, J.; Contu, R.; Cossu, L.; Latronico, M.V.; Peterson, K.L.; Indolfi, C.; Catalucci, D.; Chen, J.; et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009, 16, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Torella, D.; Iaconetti, C.; Catalucci, D.; Ellison, G.M.; Leone, A.; Waring, C.D.; Bochicchio, A.; Vicinanza, C.; Aquila, I.; Curcio, A.; et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ. Res. 2011, 109, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zheng, C.; Ye, H.; Teng, Y.; Zheng, B.; Yang, X.; Zhang, J. MicroRNA-365 inhibits vascular smooth muscle cell proliferation through targeting cyclin D1. Int. J. Med. Sci. 2014, 11, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Ham, O.; Lee, S.Y.; Choi, E.; Lee, C.Y.; Park, J.H.; Lee, J.; Seo, H.H.; Seung, M.; Min, P.K.; et al. MicroRNA-365 inhibits the proliferation of vascular smooth muscle cells by targeting cyclin D1. J. Cell Biochem. 2014, 115, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Leeper, N.J.; Raiesdana, A.; Kojima, Y.; Chun, H.J.; Azuma, J.; Maegdefessel, L.; Kundu, R.K.; Quertermous, T.; Tsao, P.S.; Spin, J.M. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J. Cell Physiol. 2011, 226, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Li, H.; Du, Y.; Shi, Q.; Zhao, L. Downregulation of microRNA-34b is responsible for the elevation of blood pressure in spontaneously hypertensive rats. Mol. Med. Rep. 2017, 15, 1031–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacurari, M.; Tchounwou, P.B. Role of microRNAs in renin-angiotensin-aldosterone system-mediated cardiovascular inflammation and remodeling. Int. J. Inflamm. 2015, 2015, 101527. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Satoh, M.; Minami, Y.; Tabuchi, T.; Itoh, T.; Nakamura, M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: Effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin. Sci. 2010, 119, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Jeunemaitre, X. Genetics of the human renin angiotensin system. J. Mol. Med. 2008, 86, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Xu, R.; Yu, H.M.; Chang, Q.; Zhong, J.C. The ACE2/apelin signaling, microRNAs, and hypertension. Int. J. Hypertens. 2015, 2015, 896861. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.R.; Unal, H.; Desnoyer, R.; Yue, H.; Bhatnagar, A.; Karnik, S.S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014, 75, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Ceolotto, G.; Papparella, I.; Bortoluzzi, A.; Strapazzon, G.; Ragazzo, F.; Bratti, P.; Fabricio, A.S.; Squarcina, E.; Gion, M.; Palatini, P.; et al. Interplay between miR-155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives. Am. J. Hypertens. 2011, 24, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Nossent, A.Y.; Eskildsen, T.V.; Andersen, L.B.; Bie, P.; Brønnum, H.; Schneider, M.; Andersen, D.C.; Welten, S.M.; Jeppesen, P.L.; Hamming, J.F.; et al. The 14q32 microRNA-487b targets the antiapoptotic insulin receptor substrate 1 in hypertension-induced remodeling of the aorta. Ann. Surg. 2013, 258, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.L.; Marques, F.Z.; Watson, A.M.; Palma-Rigo, K.; Nguyen-Huu, T.P.; Morris, B.J.; Charchar, F.J.; Davern, P.J.; Head, G.A. A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2J genetically hypertensive mice. Hypertension 2013, 62, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.J.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Romaine, S.P.R.; Denniff, M.; Eales, J.; Dormer, J.; Garrelds, I.M.; Wojnar, L.; Musialik, K.; Duda-Raszewska, B.; Kiszka, B.; et al. Signatures of miR-181a on renal transcriptome and blood pressure. Mol. Med. 2015, 21, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Winklewski, P.J.; Radkowski, M.; Wszedybyl-Winklewska, M.; Demkow, U. Brain inflammation and hypertension: The chicken or the egg? J. Neuroinflamm. 2015, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Sivasinprasasn, S.; Pantan, R.; Thummayot, S.; Tocharus, J.; Suksamrarn, A.; Tocharus, C. Cyanidin-3-glucoside attenuates angiotensin II-induced oxidative stress and inflammation in vascular endothelial cells. Chem. Biol. Interact. 2016, 260, 67–74. [Google Scholar] [CrossRef] [PubMed]
- DeCicco, D.; Zhu, H.; Brureau, A.; Schwaber, J.S.; Vadigepalli, R. MicroRNA network changes in the brain stem underlie the development of hypertension. Physiol. Genomics 2015, 47, 388–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friese, R.S.; Altshuler, A.E.; Zhang, K.; Miramontes-Gonzalez, J.P.; Hightower, C.M.; Jirout, M.L.; Salem, R.M.; Gayen, J.R.; Mahapatra, N.R.; Biswas, N.; et al. MicroRNA-22 and promoter motif polymorphisms at the Chga locus in genetic hypertension: Functional and therapeutic implications for gene expression and the pathogenesis of hypertension. Hum. Mol. Genet. 2013, 22, 3624–3640. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, S.; Ji-yan, C.; Huang, C.; Li, J.; Cai, A.; Feng, Y. Circulating miR-92a expression level in patients with essential hypertension: A potential marker of atherosclerosis. J. Hum. Hypertens. 2017, 31, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, S.; Huang, C.; Chen, J.; Li, J.; Cai, A.; Feng, Y. Circulating miRNA29 family expression levels in patients with essential hypertension as potential markers for left ventricular hypertrophy. Clin. Exp. Hypertens. 2017, 39, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Parthenakis, F.I.; Maragkoudakis, S.; Zacharis, E.A.; Petousis, S.; Kochiadakis, G.E.; Vardas, P.E. Hypertrophic and antihypertrophic microRNA levels in peripheral blood mononuclear cells and their relationship to left ventricular hypertrophy in patients with essential hypertension. J. Am. Soc. Hypertens. 2015, 9, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, J.; Zhang, W.; Chen, Y.; Zhang, K.; Popescu, L.M.; Ma, X.; Bond Lau, W.; Rong, R.; Yu, X.; et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation 2011, 124, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Hao, Y.; Chong, X.; Zhong, W. Overexpression of microRNA-506-3p aggravates the injury of vascular endothelial cells in patients with hypertension by downregulating Beclin1 expression. Exp. Ther. Med. 2018, 15, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Venardos, K.; Jones, E.; Morris, B.J.; Chin-Dusting, J.; Kaye, D.M. Identification of a novel polymorphism in the 3’UTR of the L-arginine transporter gene SLC7A1: Contribution to hypertension and endothelial dysfunction. Circulation 2007, 115, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Wu, C.; Khan, A.M.; Bloch, D.B.; Davis-Dusenbery, B.N.; Ghorbani, A.; Spagnolli, E.; Martinez, A.; Ryan, A.; Tainsh, L.T.; et al. Atrial natriuretic peptide is negatively regulated by microRNA-425. J. Clin. Investig. 2013, 123, 3378–3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maharjan, S.; Mopidevi, B.; Kaw, M.K.; Puri, N.; Kumar, A. Human aldosterone synthase gene polymorphism promotes miRNA binding and regulates gene expression. Physiol. Genomics 2014, 46, 860–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korkor, M.T.; Meng, F.B.; Xing, S.Y.; Zhang, M.C.; Guo, J.R.; Zhu, X.X.; Yang, P. Microarray analysis of differential gene expression profile in peripheral blood cells of patients with human essential hypertension. Int. J. Med. Sci. 2011, 8, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Whiting, D.R.; Guariguata, L.; Weil, C.; Shaw, J. IDF diabetes atlas: Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res. Clin. Pract. 2011, 94, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Prentki, M.; Nolan, C.J. Islet cell failure in type 2 diabetes. J. Clin. Investig. 2006, 116, 1802–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stępień, E.; Stankiewicz, E.; Zalewski, J.; Godlewski, J.; Zmudka, K.; Wybrańska, I. Number of microparticles generated during acute myocardial infarction and stable angina correlates with platelet activation. Arch. Med. Res. 2012, 43, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Stępień, E.L.; Durak-Kozica, M.; Kamińska, A.; Targosz-Korecka, M.; Libera, M.; Tylko, G.; Opalińska, A.; Kapusta, M.; Solnica, B.; Georgescu, A.; et al. Circulating ectosomes: Determination of angiogenic microRNAs in type 2 diabetes. Theranostics 2018, 8, 3874–3890. [Google Scholar] [CrossRef] [PubMed]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc. Res. 2012, 93, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Murach, K.A.; McCarthy, J.J. MicroRNAs, heart failure, and aging: Potential interactions with skeletal muscle. Heart Fail. Rev. 2017, 22, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Drummond, M.J.; McCarthy, J.J.; Fry, C.S.; Esser, K.A.; Rasmussen, B.B. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1333–E1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Baroukh, N.; Ravier, M.A.; Loder, M.K.; Hill, E.V.; Bounacer, A.; Scharfmann, R.; Rutter, G.A.; Van Obberghen, E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J. Biol. Chem. 2007, 282, 19575–19588. [Google Scholar] [CrossRef] [PubMed]
- Lovis, P.; Gattesco, S.; Regazzi, R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 2008, 389, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Muniappan, L.; Tang, G.; Ozcan, S. Identification of glucose-regulated miRNAs from pancreatic {β} cells reveals a role for miR-30d in insulin transcription. RNA 2009, 15, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbagallo, D.; Piro, S.; Condorelli, A.G.; Mascali, L.G.; Urbano, F.; Parrinello, N.; Monello, A.; Statello, L.; Ragusa, M.; Rabuazzo, A.M.; et al. miR-296-3p, miR-298-5p and their downstream networks are causally involved in the higher resistance of mammalian pancreatic α cells to cytokine-induced apoptosis as compared to β cells. BMC Genomics 2013, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef] [PubMed]
- El Ouaamari, A.; Baroukh, N.; Martens, G.A.; Lebrun, P.; Pipeleers, D.; Van Obberghen, E. miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic β-cells. Diabetes 2008, 57, 2708–2717. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Plaisance, V.; Abderrahmani, A.; Perret-Menoud, V.; Jacquemin, P.; Lemaigre, F.; Regazzi, R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006, 281, 26932–26942. [Google Scholar] [CrossRef] [PubMed]
- Herrera, B.M.; Lockstone, H.E.; Taylor, J.M.; Ria, M.; Barrett, A.; Collins, S.; Kaisaki, P.; Argoud, K.; Fernandez, C.; Travers, M.E.; et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia 2010, 53, 1099–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.A.; Wei, Y.; Sowers, J.R. Role of mitochondrial dysfunction in insulin resistance. Circ. Res. 2008, 102, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Prabu, P.; Rome, S.; Sathishkumar, C.; Aravind, S.; Mahalingam, B.; Shanthirani, C.S.; Gastebois, C.; Villard, A.; Mohan, V.; Balasubramanyam, M. Circulating miRNAs of “asian indian phenotype” identified in subjects with impaired glucose tolerance and patients with type 2 diabetes. PLoS ONE 2015, 10, e0128372. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Sci. 2013, 126, 2678–2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Yang, Z.M.; Chen, L.H.; Hong, M.; Chen, Y.Y.; Yang, X.R.; Tang, S.M.; Yuan, Q.F.; Chen, W.W. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol. Med. Rep. 2017, 15, 2143–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, H.S.; Park, S.Y.; Ma, D.; Zhang, J.; Lee, W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE 2011, 6, e17343. [Google Scholar] [CrossRef]
- Fernandez-Twinn, D.S.; Alfaradhi, M.Z.; Martin-Gronert, M.S.; Duque-Guimaraes, D.E.; Piekarz, A.; Ferland-McCollough, D.; Bushell, M.; Ozanne, S.E. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol. Metab. 2014, 3, 325–333. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, Y.; Yang, X.; He, X.; Zhang, H.; Zhang, L. The feedback regulation of PI3K-miR-19a, and MAPK-miR-23b/27b in endothelial cells under shear stress. Molecules 2012, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, J.H.; Ha, M.; Nam, J.W.; Kim, V.N. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat. Struct. Mol. Biol. 2009, 16, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Lin, C.; Ren, J.; Liu, J. MicroRNA-384-5p regulates ischemia-induced cardioprotection by targeting phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta (PI3K p110δ). Apoptosis 2013, 18, 260–270. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Zhu, L.; Gupta, N.; Chang, Y.; Fang, F. Overexpression of Micro Ribonucleic Acid 29, Highly Up-Regulated in Diabetic Rats, Leads to Insulin Resistance in 3T3-L1 Adipocytes. Mol. Endocrinol. 2007, 21, 2785–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dooley, J.; Garcia-Perez, J.E.; Sreenivasan, J.; Schlenner, S.M.; Vangoitsenhoven, R.; Papadopoulou, A.S.; Tian, L.; Schonefeldt, S.; Serneels, L.; Deroose, C.; et al. The microRNA-29 family dictates the balance between homeostatic and pathological glucose handling in diabetes and obesity. Diabetes 2016, 65, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Vatandoost, N.; Amini, M.; Iraj, B.; Momenzadeh, S.; Salehi, R. Dysregulated miR-103 and miR-143 expression in peripheral blood mononuclear cells from induced prediabetes and type 2 diabetes rats. Gene 2015, 572, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Feinberg, M.W.; Hull, S.; Kuo, C.T.; Watanabe, M.; Banerjee, S.S.; DePina, A.; Haspel, R.; Jain, M.K. The Krüppel-like Factor KLF15 Regulates the Insulin-sensitive Glucose Transporter GLUT4. J. Biol. Chem. 2002, 277, 34322–34328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, T.; Ono, K.; Nishi, H.; Iwanaga, Y.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Takanabe, R.; Hasegawa, K.; Kita, T.; et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem. Biophys. Res. Commun. 2009, 389, 315–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, H.Y.; Hu, B.; Hu, X.B.; Zhong, J.; Feng, S.D.; Qin, L.; Liu, G.; Wen, G.B.; Liao, D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes 2012, 120, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Buchan, R.J.; Cook, S.A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 2010, 86, 410–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.; Meng, X.; Che, H.; Shen, N.; Xiao, D.; Song, X.; Liang, M.; Fu, X.; Ju, J.; Li, Y.; et al. Regulation of insulin resistance by multiple miRNAs via targeting the GLUT4 signalling pathway. Cell. Physiol. Biochem. 2016, 38, 2063–2078. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, G.; Yang, C.; Zhou, K.; Shen, B.; Liang, H.; Jiang, X. The role of circulating microRNA-126 (miR-126): A novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int. J. Mol. Sci. 2014, 15, 10567–10577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, L.; Shang, Q.; Lv, C.; Wang, C.; Su, B. Circulating miR-126 is a potential biomarker to predict the onset of type 2 diabetes mellitus in susceptible individuals. Biochem. Biophys. Res. Commun. 2015, 463, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Spazzafumo, L.; Bonafè, M.; Recchioni, R.; Prattichizzo, F.; Marcheselli, F.; Micolucci, L.; Mensà, E.; Giuliani, A.; Santini, G.; et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget 2015, 6, 35372–35382. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Rovira, O.; Guerra, E.; Esteve, E.; Xifra, G.; Martínez, C.; Ricart, W.; Rieusset, J.; et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014, 37, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Cao, J.T.; Zhang, B.; Zhou, Q.; Shen, C.X.; Wang, C.Q. Downregulation of microRNA-126 in endothelial progenitor cells from diabetes patients, impairs their functional properties, via target gene Spred-1. J. Mol. Cell. Cardiol. 2012, 53, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiang, X.; Ma, H.; Wang, Y.; Xue, P.; Liu, Y. SIRT1 and insulin resistance. J. Diabetes Complicat. 2016, 30, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, C.; Qi, W.; Zhang, Y.; Zhang, F.; Wu, J.X.; Hu, Y.N.; Wu, D.M.; Liu, Y.; Yan, T.T.; et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012, 55, 2032–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raut, S.K.; Singh, G.B.; Rastogi, B.; Saikia, U.N.; Mittal, A.; Dogra, N.; Singh, S.; Prasad, R.; Khullar, M. miR-30c and miR-181a synergistically modulate p53-p21 pathway in diabetes induced cardiac hypertrophy. Mol. Cell. Biochem. 2016, 417, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Zhu, M.; Mao, X.; Long, M.; Du, X.; Wu, Y.; Abudureyimu, K.; Zhang, C.; Wang, Y.; Tao, Y.; et al. MicroRNA-130a expression is decreased in Xinjiang Uygur patients with type 2 diabetes mellitus. Am. J. Transl. Res. 2015, 7, 1984–1991. [Google Scholar] [PubMed]
- Yan, S.; Wang, T.; Huang, S.; Di, Y.; Huang, Y.; Liu, X.; Luo, Z.; Han, W.; An, B. Differential expression of microRNAs in plasma of patients with prediabetes and newly diagnosed type 2 diabetes. Acta Diabetol. 2016, 53, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Pescador, N.; Pérez-Barba, M.; Ibarra, J.M.; Corbatón, A.; Martínez-Larrad, M.T.; Serrano-Ríos, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 2007, 8, 657–662. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M.I. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010, 363, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Lin, Y.; Zhang, Y.; Yang, J.; Zhang, Y.; Liu, H.; Zhang, B. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: A large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med. Genet. 2009, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Jurado, J.; Ybarra, J.; Romeo, J.H.; Garcia, M.; Zabaleta-Del-Olmo, E. Angiotensin-converting enzyme gene single polymorphism as a genetic biomarker of diabetic peripheral neuropathy: Longitudinal prospective study. J. Diabetes Complicat. 2012, 26, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.Y.; Li, H.F.; Lei, Y.; Rao, Y.W.; Tan, Z.X.; Liu, H.J.; Yao, G.D.; Hou, B.; Sun, M.L. Association of adiponectin gene polymorphisms with an elevated risk of diabetic peripheral neuropathy in type 2 diabetes patients. J. Diabetes Complicat. 2015, 29, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Chanprasertyothin, S.; Jongjaroenprasert, W.; Ongphiphadhanakul, B. The association of soluble IGF2R and IGF2R gene polymorphism with type 2 diabetes. J. Diabetes Res. 2015, 2015, 216383. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Nafis, S.; Kalaiarasan, P.; Rai, E.; Sharma, S.; Bamezai, R.N. Understanding genetic heterogeneity in type 2 diabetes by delineating physiological phenotypes: SIRT1 and its gene network in impaired insulin secretion. Rev. Diabet. Stud. 2016, 13, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.I.G.; Simpson, H.L.; Sönksen, P.H. The role of the growth hormone-insulin-like growth factor axis in glucose homeostasis. Diabet. Med. 2003, 20, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.K.; Dunger, D.B. Developmental aspects in the pathogenesis of type 2 diabetes. Mol. Cell. Endocrinol. 2001, 185, 145–149. [Google Scholar] [CrossRef]
- McCann, J.A.; Xu, Y.Q.; Frechette, R.; Guazzarotti, L.; Polychronakos, C. The insulin-like growth factor-II receptor gene is associated with type 1 diabetes: Evidence of a maternal effect. J. Clin. Endocrinol. Metab. 2004, 89, 5700–5706. [Google Scholar] [CrossRef] [PubMed]
- Villuendas, G.; Botella-Carretero, J.I.; López-Bermejo, A.; Gubern, C.; Ricart, W.; Fernández-Real, J.M.; San Millán, J.L.; Escobar-Morreale, H.F. The ACAA-insertion/deletion polymorphism at the 3’ UTR of the IGF-II receptor gene is associated with type 2 diabetes and surrogate markers of insulin resistance. Eur. J. Endocrinol. 2006, 155, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Lv, K.; Guo, Y.; Zhang, Y.; Wang, K.; Jia, Y.; Sun, S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem. Biophys. Res. Commun. 2008, 374, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Hatefi, Z.; Soltani, G.; Khosravi, S.; Kazemi, M.; Salehi, A.R.; Salehi, R. Micro R-410 binding site single nucleotide polymorphism rs13702 in lipoprotein lipase gene is effective to increase susceptibility to type 2 diabetes in iranian population. Adv. Biomed. Res. 2018, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Lovis, P.; Roggli, E.; Laybutt, D.R.; Gattesco, S.; Yang, J.Y.; Widmann, C.; Abderrahmani, A.; Regazzi, R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 2008, 57, 2728–2736. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.J.A.; Olson, E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc. Natl. Acad. Sci. USA 2011, 108, 21075–21080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Peng, R.; Li, T.; Luo, X.; Peng, H.; Zha, H.; Yin, P.; Wen, L.; Zhang, Z. A potentially functional polymorphism in the regulatory region of let-7a-2 is associated with an increased risk for diabetic nephropathy. Gene 2013, 527, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Chen, Y.J.; Sun, L.L.; Zhang, S.J.; Zhou, Z.Y.; Qiao, H. Affection of single-nucleotide polymorphisms in miR-27a, miR-124a, and miR-146a on susceptibility to type 2 diabetes mellitus in Chinese Han people. Chin. Med. J. 2015, 128, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.D.; Ryan, D.H.; Apovian, C.M.; Ard, J.D.; Comuzzie, A.G.; Donato, K.A.; Hu, F.B.; Hubbard, V.S.; Jakicic, J.M.; Kushner, R.F.; et al. 2013 AHA/ACC/TOS Guideline for the Management of Overweight and Obesity in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and The Obesity Society. J. Am. Coll. Cardiol. 2014, 63, 2985–3023. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Huang, F.; Gu, X.; Zhang, M.; Wen, J.; Wang, X.; You, L.; Cui, X.; Ji, C.; Guo, X. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 2016, 7, 40830–40845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.Z.; Scheideler, M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Xu, G.; Ji, C.; Shi, C.; Shen, Y.; Chen, L.; Zhu, L.; Yang, L.; Zhao, Y.; Guo, X. The role of microRNA-26b in human adipocyte differentiation and proliferation. Gene 2014, 533, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ji, C.; Song, G.; Zhao, C.; Shi, C.; Song, L.; Chen, L.; Yang, L.; Huang, F.; Pang, L.; et al. MiR-26b modulates insulin sensitivity in adipocytes by interrupting the PTEN/PI3K/AKT pathway. Int. J. Obes. 2015, 39, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Skårn, M.; Namløs, H.M.; Noordhuis, P.; Wang, M.Y.; Meza-Zepeda, L.A.; Myklebost, O. Adipocyte differentiation of human bone marrow-derived stromal cells is modulated by microRNA-155, microRNA-221, and microRNA-222. Stem Cells Dev. 2012, 21, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Hamam, D.; Ali, D.; Vishnubalaji, R.; Hamam, R.; Al-Nbaheen, M.; Chen, L.; Kassem, M.; Aldahmash, A.; Alajez, N.M. microRNA-320/RUNX2 axis regulates adipocytic differentiation of human mesenchymal (skeletal) stem cells. Cell Death Dis. 2014, 5, e1499. [Google Scholar] [CrossRef] [PubMed]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 Family Is Required for Human Adipogenesis and Drives Characteristics of Brown Adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, A.; Barber, T.M.; de Bunt, M.; Rudge, S.A.; Zhang, Q.; Lachlan, K.L.; Cooper, N.S.; Linden, H.; Levy, J.C.; Wakelam, M.J.; et al. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N. Engl. J. Med. 2012, 367, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Togliatto, G.; Dentelli, P.; Gili, M.; Gallo, S.; Deregibus, C.; Biglieri, E.; Iavello, A.; Santini, E.; Rossi, C.; Solini, A.; et al. Obesity reduces the pro-angiogenic potential of adipose tissue stem cell-derived extracellular vesicles (EVs) by impairing miR-126 content: Impact on clinical applications. Int. J. Obes. 2016, 40, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Jansen, F.; Yang, X.; Hoelscher, M.; Cattelan, A.; Schmitz, T.; Proebsting, S.; Wenzel, D.; Vosen, S.; Franklin, B.S.; Fleischmann, B.K.; et al. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation 2013, 128, 2026–2038. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Hwang, S.J.; Bae, Y.C.; Jung, J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells 2009, 27, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Fischer, A.; Muhly-Reinholz, M.; Zeiher, A.M.; Dimmeler, S. Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity (Silver Spring) 2014, 22, 2352–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, L.; Chen, Y.; Niu, Y.; Chen, W.; Wang, Q.; Xiao, S.; Li, A.; Xie, Y.; Li, J.; Zhao, X.; et al. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics 2010, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Roldan, M.; Macias-Gonzalez, M.; Garcia, R.; Tinahones, F.J.; Martin, M. Obesity short-circuits stemness gene network in human adipose multipotent stem cells. FASEB J. 2011, 25, 4111–4126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Dai, Y.M.; Ji, C.B.; Yang, L.; Shi, C.M.; Xu, G.F.; Pang, L.X.; Huang, F.Y.; Zhang, C.M.; Guo, X.R. MiR-146b is a regulator of human visceral preadipocyte proliferation and differentiation and its expression is altered in human obesity. Mol. Cell. Endocrinol. 2014, 393, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Tanaka, Y.; Kawamori, R.; Maeda, S. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic β-cell line. Mol. Endocrinol. 2006, 20, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Condorelli, A.G.; Piro, S.; Parrinello, N.; Fløyel, T.; Ragusa, M.; Rabuazzo, A.M.; Størling, J.; Purrello, F.; Di Pietro, C.; et al. CEBPA exerts a specific and biologically important proapoptotic role in pancreatic β cells through its downstream network targets. Mol. Biol. Cell 2014, 25, 2333–2341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanazawa, A.; Kawamura, Y.; Sekine, A.; Iida, A.; Tsunoda, T.; Kashiwagi, A.; Tanaka, Y.; Babazono, T.; Matsuda, M.; Kawai, K.; et al. Single nucleotide polymorphisms in the gene encoding Krüppel-like factor 7 are associated with type 2 diabetes. Diabetologia 2005, 48, 1315–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinelli, R.; Nardelli, C.; Pilone, V.; Buonomo, T.; Liguori, R.; Castanò, I.; Buono, P.; Masone, S.; Persico, G.; Forestieri, P.; et al. miR-519d Overexpression Is Associated with Human Obesity. Obesity 2010, 18, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Sinnaeve, P.; der Schueren, B.; Mathieu, C.; Janssens, S.; Holvoet, P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J. Clin. Endocrinol. Metab. 2012, 97, E1213–E1218. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A.; Syed, R.; Duggineni, D.; Rutsky, J.; Rengasamy, P.; Zhang, J.; Huang, K.; Needleman, B.; Mikami, D.; Perry, K.; et al. Visceral Adipose MicroRNA 223 Is Upregulated in Human and Murine Obesity and Modulates the Inflammatory Phenotype of Macrophages. PLoS ONE 2016, 11, e0165962. [Google Scholar] [CrossRef] [PubMed]
- Iacomino, G.; Russo, P.; Stillitano, I.; Lauria, F.; Marena, P.; Ahrens, W.; de Luca, P.; Siani, A. Circulating microRNAs are deregulated in overweight/obese children: Preliminary results of the I.Family study. Genes Nutr. 2016, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Dai, X.; Zhan, J.; Zhang, Y.; Zhang, H.; Zhang, H.; Zeng, S.; Xi, W. Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS 2015, 123, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Mercader, J.M.; Catalán, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gómez-Ambrosi, J.; Anglada, R.; Fernández-Formoso, J.A.; Ricart, W.; et al. Targeting the circulating microRNA signature of obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Qiao, P.; Wang, L. Circulating microRNA-223 as a potential biomarker for obesity. Obes. Res. Clin. Pract. 2015, 9, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Kilic, I.D.; Dodurga, Y.; Uludag, B.; Alihanoglu, Y.I.; Yildiz, B.S.; Enli, Y.; Secme, M.; Bostancı, H.E. MicroRNA-143 and -223 in obesity. Gene 2015, 560, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Bruun, J.M.; Stallknecht, B.; Helge, J.W.; Richelsen, B. Interleukin-18 in plasma and adipose tissue: Effects of obesity, insulin resistance, and weight loss. Eur. J. Endocrinol. 2007, 157, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Wen, H.; Ting, J.P.Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Kaplanski, G. Interleukin-18: Biological properties and role in disease pathogenesis. Immunol. Rev. 2018, 281, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.; Li, Z.; Wu, Y.; Zhang, C.; Ai, X.; Wang, C.; Shi, H.; Hui, M.; Xie, B.; et al. Cross talk between vascular smooth muscle cells and monocytes through interleukin-1β/interleukin-18 signaling promotes vein graft thickening. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- Sims, J.E. IL-1 and IL-18 receptors, and their extended family. Curr. Opin. Immunol. 2002, 14, 117–122. [Google Scholar] [CrossRef]
- Martínez-Barquero, V.; Marco, G.; de Martínez-Hervas, S.; Adam-Felici, V.; Pérez-Soriano, C.; Gonzalez-Albert, V.; Rojo, G.; Ascaso, J.F.; Real, J.T.; Garcia-Garcia, A.B.; et al. Are IL18RAP gene polymorphisms associated with body mass regulation? A cross-sectional study. BMJ Open 2017, 7, e017875. [Google Scholar] [CrossRef] [PubMed]
- Zaragosi, L.E.; Wdziekonski, B.; Brigand, K.L.; Villageois, P.; Mari, B.; Waldmann, R.; Dani, C.; Barbry, P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011, 12, R64. [Google Scholar] [CrossRef] [PubMed]
- Meale, S.J.; Romao, J.M.; He, M.L.; Chaves, A.V.; McAllister, T.A.; Guan, L.L. Effect of diet on microRNA expression in ovine subcutaneous and visceral adipose tissues. J. Anim. Sci. 2014, 92, 3328–3337. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, M.; Gomez-Valadés, A.G.; Ramirez, S.; Gomis, R.; Claret, M. Hypothalamic miRNAs: Emerging roles in energy balance control. Front. Neurosci. 2015, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Richardson, K.; Lai, C.Q.; Parnell, L.D.; Lee, Y.C.; Ordovas, J.M. A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS. BMC Genomics 2011, 12, 504. [Google Scholar] [CrossRef] [PubMed]
- Tansey, J.T.; Sztalryd, C.; Gruia-Gray, J.; Roush, D.L.; Zee, J.V.; Gavrilova, O.; Reitman, M.L.; Deng, C.X.; Li, C.; Kimmel, A.R.; et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA 2001, 98, 6494–6499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcântara, V.M.; Oliveira, L.C.; Réa, R.R.; Suplicy, H.L.; Chautard-Freire-Maia, E.A. Butyrylcholinesterase activity and metabolic syndrome in obese patients. Clin. Chem. Lab. Med. 2005, 43, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Sisková, K.; Bilka, F.; Adameová, A.; Balazová, A.; Mydla, M.; Pauliková, I. Influence of lipid imbalance on butyrylcholinesterase activity and biotransformation efficiency. Pharmazie 2012, 67, 345–350. [Google Scholar] [PubMed]
- Lima, J.K.; Leite, N.; Turek, L.V.; Souza, R.L.R.; da Silva Timossi, L.; Osiecki, A.C.V.; Osiecki, R.; Furtado-Alle, L. 1914G variant of BCHE gene associated with enzyme activity, obesity and triglyceride levels. Gene 2013, 532, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Duygu, B.; Da Costa Martins, P.A. miR-21: A star player in cardiac hypertrophy. Cardiovasc. Res. 2015, 105, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Sell, H.; Dietze-Schroeder, D.; Kaiser, U.; Eckel, J. Monocyte chemotactic protein-1 is a potential player in the negative cross-talk between adipose tissue and skeletal muscle. Endocrinology 2006, 147, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Huang, H.; Xu, Y.; Zhu, H.; Zhong, C. MiR-222 in cardiovascular diseases: Physiology and pathology. BioMed Res. Int. 2017, 2017, 4962426. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Fan, J.; Chen, N. A novel regulator of type II diabetes: MicroRNA-143. Trends Endocrinol. Metab. 2018, 29, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Ono, K. MicroRNA links obesity and impaired glucose metabolism. Cell Res. 2011, 21, 864–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.T.; Wong, M.T.K.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA Profiles in Patients with Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef] [PubMed]
- Oja, P.; Titze, S. Physical activity recommendations for public health: Development and policy context. EPMA J. 2011, 2, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, L.; Yang, Y.J.; Ge, R.K.; Zhou, M.; Hu, H.; Liu, H.; Cui, J.; Li, L.L.; Dong, Y.F.; et al. Aerobic exercise improves endothelial function and serum adropin levels in obese adolescents independent of body weight loss. Sci. Rep. 2017, 7, 17717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clauss, S.; Wakili, R.; Hildebrand, B.; Kääb, S.; Hoster, E.; Klier, I.; Martens, E.; Hanley, A.; Hanssen, H.; Halle, M.; et al. MicroRNAs as biomarkers for acute atrial remodeling in marathon runners (the mirathon study—A sub-study of the munich marathon study). PLoS ONE 2016, 11, e0148599. [Google Scholar] [CrossRef] [PubMed]
- Börjesson, M.; Onerup, A.; Lundqvist, S.; Dahlöf, B. Physical activity and exercise lower blood pressure in individuals with hypertension: Narrative review of 27 RCTs. Br. J. Sports Med. 2016, 50, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.R.; O’Sullivan, A.J.; Fiatarone Singh, M.A. Exercise or physical activity and cognitive function in adults with type 2 diabetes, insulin resistance or impaired glucose tolerance: A systematic review. Eur. Rev. Aging Phys. Act. 2018, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Ingul, C.B.; Dias, K.A.; Tjonna, A.E.; Follestad, T.; Hosseini, M.S.; Timilsina, A.S.; Hollekim-Strand, S.M.; Ro, T.B.; Davies, P.S.W.; Cain, P.A.; et al. Effect of High Intensity Interval Training on Cardiac Function in Children with Obesity: A Randomised Controlled Trial. Prog. Cardiovasc. Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Junior, S.J.; Sá, C.G.A.d.B.; Rodrigues, P.A.F.; Oliveira, A.J.; Fernandes-Filho, J. Physical exercise and morbid obesity: A systematic review. Arq. Bras. Cir. Dig. 2013, 26, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Ferioli, M.; Zauli, G.; Martelli, A.M.; Vitale, M.; McCubrey, J.A.; Ultimo, S.; Capitani, S.; Neri, L.M. Impact of physical exercise in cancer survivors during and after antineoplastic treatments. Oncotarget 2018, 9, 14005–14034. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ai, D.; Zhang, N. Exercise benefits coronary heart disease. Adv. Exp. Med. Biol. 2017, 1000, 3–7. [Google Scholar] [CrossRef] [PubMed]
- do Prado, D.M.L.; Rocco, E.A. The benefits of exercise training on aerobic capacity in patients with heart failure and preserved ejection fraction. Adv. Exp. Med. Biol. 2017, 1000, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Soci, U.P.; Oliveira, E.M. Eccentric and concentric cardiac hypertrophy induced by exercise training: MicroRNAs and molecular determinants. Braz. J. Med. Biol. Res. 2011, 44, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Konhilas, J.P.; Watson, P.A.; Maass, A.; Boucek, D.M.; Horn, T.; Stauffer, B.L.; Luckey, S.W.; Rosenberg, P.; Leinwand, L.A. Exercise can prevent and reverse the severity of hypertrophic cardiomyopathy. Circ. Res. 2006, 98, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Novoa, U.; Arauna, D.; Moran, M.; Nuñez, M.; Zagmutt, S.; Saldivia, S.; Valdes, C.; Villaseñor, J.; Zambrano, C.G.; Gonzalez, D.R. High-intensity exercise reduces cardiac fibrosis and hypertrophy but does not restore the nitroso-redox imbalance in diabetic cardiomyopathy. Oxid. Med. Cell. Longev. 2017, 2017, 7921363. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, P.; Temporelli, P.L.; Corrà, U.; Tavazzi, L.; ELVD-CHF Study Group. Antiremodeling effect of long-term exercise training in patients with stable chronic heart failure: Results of the exercise in left ventricular dysfunction and chronic heart failure (ELVD-CHF) trial. Circulation 2003, 108, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wan, W.; Powers, A.S.; Li, J.; Ji, L.L.; Lao, S.; Wilson, B.; Erikson, J.M.; Zhang, J.Q. Effects of exercise training on cardiac function and myocardial remodeling in post myocardial infarction rats. J. Mol. Cell. Cardiol. 2008, 44, 114–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.; Åkerström, T.; Rinnov, A.; Yfanti, C.; Scheele, C.; Pedersen, B.K.; Laye, M.J. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS ONE 2014, 9, e87308. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.J.J.; Bye, A.; el Azzouzi, H.; Wisløff, U. MicroRNAs as important regulators of exercise adaptation. Prog. Cardiovasc. Dis. 2017, 60, 130–151. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Qi, J.; Meng, S.; Wen, B.; Zhang, J. Swimming exercise training-induced left ventricular hypertrophy involves microRNAs and synergistic regulation of the PI3K/AKT/mTOR signaling pathway. Eur. J. Appl. Physiol. 2013, 113, 2473–2486. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, Z. Swimming training affects apoptosis-related microRNAs and reduces cardiac apoptosis in mice. Gene Physiol. Biophys. 2016, 35, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.G.; Silva, M.S.M.; Duarte, N.E.; Bolani, W.; Alves, C.R.; Junior, J.R.L.; da Silva, J.L.; de Oliveira, P.A.; Alves, G.B.; de Oliveira, E.M.; et al. PBMCs express a transcriptome signature predictor of oxygen uptake responsiveness to endurance exercise training in men. Physiol. Genomics 2015, 47, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radom-Aizik, S.; Zaldivar, F.; Leu, S.Y.; Adams, G.R.; Oliver, S.; Cooper, D.M. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin. Transl. Sci. 2012, 5, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Baggish, A.L.; Hale, A.; Weiner, R.B.; Lewis, G.D.; Systrom, D.; Wang, F.; Wang, T.J.; Chan, S.Y. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J. Physiol. 2011, 589, 3983–3994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baggish, A.L.; Park, J.; Min, P.K.; Isaacs, S.; Parker, B.A.; Thompson, P.D.; Troyanos, C.; D’Hemecourt, P.; Dyer, S.; Thiel, M.; et al. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J. Appl. Physiol. 2014, 116, 522–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Zhou, Q.; Che, L.; Das, S.; Wang, L.; Jiang, J.; Li, G.; Xu, J.; Yao, J.; Wang, H.; et al. Circulating miR-21, miR-378, and miR-940 increase in response to an acute exhaustive exercise in chronic heart failure patients. Oncotarget 2016, 7, 12414–12425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza, R.W.; Fernandez, G.J.; Cunha, J.P.; Piedade, W.P.; Soares, L.C.; Souza, P.A.; de Campos, D.H.; Okoshi, K.; Cicogna, A.C.; Dal-Pai-Silva, M.; et al. Regulation of cardiac microRNAs induced by aerobic exercise training during heart failure. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1629–H1641. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, C.; Zhang, A.; Cai, H.; Price, S.R.; Wang, X.H. MicroRNA-23a and microRNA-27a mimic exercise by ameliorating CKD-induced muscle atrophy. J. Am. Soc. Nephrol. 2017, 28, 2631–2640. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Hashimoto, N.Y.; Magalhães, F.C.; Fernandes, F.B.; Casarini, D.E.; Carmona, A.K.; Krieger, J.E.; Phillips, M.I.; Oliveira, E.M. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory MicroRNAs, decreased angiotensin-converting enzyme-angiotensin ii, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1-7). Hypertension 2011, 58, 182–189. [Google Scholar] [CrossRef] [PubMed]
- da Silva, N.D.; Fernandes, T.; Soci, U.P.R.; Monteiro, A.W.A.; Phillips, M.I.; de Oliveira, E.M. Swimming training in rats increases cardiac MicroRNA-126 expression and angiogenesis. Med. Sci. Sports Exerc. 2012, 44, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.L.; Fernandes, T.; Soci, U.P.; Silveira, A.C.; Barretti, D.L.; Negrão, C.E.; Oliveira, E.M. Obesity downregulates microRNA-126 inducing capillary rarefaction in skeletal muscle: Effects of aerobic exercise training. Oxid. Med. Cell. Longev. 2017, 2017, 2415246. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F.; Haddad, F.; Cooper, D.M. Impact of brief exercise on peripheral blood NK cell gene and microRNA expression in young adults. J. Appl. Physiol. 2013, 114, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Uhlemann, M.; Möbius-Winkler, S.; Fikenzer, S.; Adam, J.; Redlich, M.; Möhlenkamp, S.; Hilberg, T.; Schuler, G.C.; Adams, V. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur. J. Prev. Cardiol. 2014, 21, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, N.C.; Cohen, C.R.; Santos, K.G.; Castro, M.A.; Biolo, A.; Frick, L.; Silvello, D.; Lopes, A.; Schneider, S.; Andrades, M.E.; et al. An analysis of the global expression of microRNAs in an experimental model of physiological left ventricular hypertrophy. PLoS ONE 2014, 9, e93271. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo-Calvo, D.; Dávalos, A.; Montero, A.; García-González, Á.; Tyshkovska, I.; González-Medina, A.; Soares, S.M.A.; Martínez-Camblor, P.; Casas-Agustench, P.; Rabadán, M.; et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J. Appl. Physiol. 2015, 119, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keidar, S.; Kaplan, M.; Gamliel-Lazarovich, A. ACE2 of the heart: From angiotensin I to angiotensin (1-7). Cardiovasc. Res. 2007, 73, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Wang, B.; Zhang, X.F.; Ma, Y.P.; Liu, J.D.; Wang, X.Z. Contribution of renin-angiotensin system to exercise-induced attenuation of aortic remodeling and improvement of endothelial function in spontaneously hypertensive rats. Cardiovasc. Pathol. 2014, 23, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Nakamuta, J.S.; Magalhães, F.C.; Roque, F.R.; Lavini-Ramos, C.; Schettert, I.T.; Coelho, V.; Krieger, J.E.; Oliveira, E.M. Exercise training restores the endothelial progenitor cells number and function in hypertension: Implications for angiogenesis. J. Hypertens. 2012, 30, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lee, A.; Wigg, J.; Peshavariya, H.; Liu, P.; Zhang, H. miR-126 regulation of angiogenesis in age-related macular degeneration in CNV mouse model. Int. J. Mol. Sci. 2016, 17, 895. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wang, F.; Shao, M.; Wang, Y.; Zhu, H. MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1. Vasc. Pharmacol. 2017, 88, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, J.; Zhu, H.; Wei, X.; Platt, C.; Damilano, F.; Xiao, C.; Bezzerides, V.; Boström, P.; Che, L.; et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015, 21, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, Y.; Wang, X.; Mu, X.; Qin, D.; Huang, W.; Alshahrani, S.; Nieman, M.; Peng, J.; Essandoh, K.; et al. Overexpression of miR-223 Tips the Balance of Pro- and Anti-hypertrophic Signaling Cascades toward Physiologic Cardiac Hypertrophy. J. Biol. Chem. 2016, 291, 15700–15713. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.D.; Zeng, K.; Liu, W.L.; Gao, Y.G.; Gong, C.S.; Zhang, C.X.; Chen, Y.Q. Effect of aerobic exercise on miRNA-TLR4 signaling in atherosclerosis. Int. J. Sports Med. 2014, 35, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Magalhães, F.C.; Roque, F.R.; Phillips, M.I.; Oliveira, E.M. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: Role of microRNAs-16, -21, and -126. Hypertension 2012, 59, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Velmurugan, G.; Shanmugha Rajan, K.; Ramprasath, T.; Kalpana, K. MiRNAs with apoptosis regulating potential are differentially expressed in chronic exercise-induced physiologically hypertrophied hearts. PLoS ONE 2015, 10, e0121401. [Google Scholar] [CrossRef] [PubMed]
- Margolis, L.M.; Rivas, D.A.; Berrone, M.; Ezzyat, Y.; Young, A.J.; McClung, J.P.; Fielding, R.A.; Pasiakos, S.M. Prolonged calorie restriction downregulates skeletal muscle mtorc1 signaling independent of dietary protein intake and associated microRNA expression. Front. Physiol. 2016, 7, 445. [Google Scholar] [CrossRef] [PubMed]
- Radom-Aizik, S.; Zaldivar, F.P.; Haddad, F.; Cooper, D.M. Impact of brief exercise on circulating monocyte gene and microRNA expression: Implications for atherosclerotic vascular disease. Brain Behav. Immun. 2014, 39, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhang, D.L.; Long, B.; An, T.; Zhang, J.; Zhou, L.Y.; Liu, C.Y.; Li, P.F. NFAT4-dependent miR-324-5p regulates mitochondrial morphology and cardiomyocyte cell death by targeting Mtfr1. Cell Death Dis. 2015, 6, e2007. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.T. Overview of microRNAs in cardiac hypertrophy, fibrosis, and apoptosis. Int. J. Mol. Sci. 2016, 17, 749. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.P.C.; Oliveira-Jr, G.P.; Madrid, B.; Almeida, J.A.; Franco, O.L.; Pereira, R.W. Circulating miR-1, miR-133a, and miR-206 levels are increased after a half-marathon run. Biomarkers 2014, 19, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Min, P.K.; Park, J.; Isaacs, S.; Taylor, B.A.; Thompson, P.D.; Troyanos, C.; D’Hemecourt, P.; Dyer, S.; Chan, S.Y.; Baggish, A.L. Influence of statins on distinct circulating microRNAs during prolonged aerobic exercise. J. Appl. Physiol. 2016, 120, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Puthanveetil, P.; Feng, B.; Matkovich, S.J.; Dorn, G.W.; Chakrabarti, S. Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. J. Cell. Mol. Med. 2014, 18, 415–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, J.; Børsheim, E.; Carvalho, E. The role of microRNAs in diabetic complications-special emphasis on wound healing. Genes 2014, 5, 926–956. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lan, H.Y.; Roukos, D.H.; Cho, W.C. Application of microRNAs in diabetes mellitus. J. Endocrinol. 2014, 222, R1–R10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Torkamandi, S.; Saffari, S.; Iranpour, M.; Omrani, M.D. Association of miR-146a expression and type 2 diabetes mellitus: A meta-analysis. Int. J. Mol. Cell. Med. 2017, 6, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Sawada, S.; Kon, M.; Wada, S.; Ushida, T.; Suzuki, K.; Akimoto, T. Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PLoS ONE 2013, 8, e70823. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-κB signaling. J. Mol. Cell. Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory microRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Güller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidsen, P.K.; Gallagher, I.J.; Hartman, J.W.; Tarnopolsky, M.A.; Dela, F.; Helge, J.W.; Timmons, J.A.; Phillips, S.M. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 2011, 110, 309–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.; Ma, F.; Li, W.; Ouyang, S.; Liu, Z.; Wu, J. miR-206-3p inhibits 3T3-L1 cell adipogenesis via the c-Met/PI3K/Akt pathway. Int. J. Mol. Sci. 2017, 18, 1510. [Google Scholar] [CrossRef] [PubMed]
- Mooren, F.C.; Viereck, J.; Krüger, K.; Thum, T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H557–H563. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Improta Caria, A.C.; Nonaka, C.K.V.; Pereira, C.S.; Soares, M.B.P.; Macambira, S.G.; Souza, B.S.d.F. Exercise Training-Induced Changes in MicroRNAs: Beneficial Regulatory Effects in Hypertension, Type 2 Diabetes, and Obesity. Int. J. Mol. Sci. 2018, 19, 3608. https://doi.org/10.3390/ijms19113608
Improta Caria AC, Nonaka CKV, Pereira CS, Soares MBP, Macambira SG, Souza BSdF. Exercise Training-Induced Changes in MicroRNAs: Beneficial Regulatory Effects in Hypertension, Type 2 Diabetes, and Obesity. International Journal of Molecular Sciences. 2018; 19(11):3608. https://doi.org/10.3390/ijms19113608
Chicago/Turabian StyleImprota Caria, Alex Cleber, Carolina Kymie Vasques Nonaka, Ciro Silveira Pereira, Milena Botelho Pereira Soares, Simone Garcia Macambira, and Bruno Solano de Freitas Souza. 2018. "Exercise Training-Induced Changes in MicroRNAs: Beneficial Regulatory Effects in Hypertension, Type 2 Diabetes, and Obesity" International Journal of Molecular Sciences 19, no. 11: 3608. https://doi.org/10.3390/ijms19113608
APA StyleImprota Caria, A. C., Nonaka, C. K. V., Pereira, C. S., Soares, M. B. P., Macambira, S. G., & Souza, B. S. d. F. (2018). Exercise Training-Induced Changes in MicroRNAs: Beneficial Regulatory Effects in Hypertension, Type 2 Diabetes, and Obesity. International Journal of Molecular Sciences, 19(11), 3608. https://doi.org/10.3390/ijms19113608