STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum
Abstract
:1. Introduction
2. Results
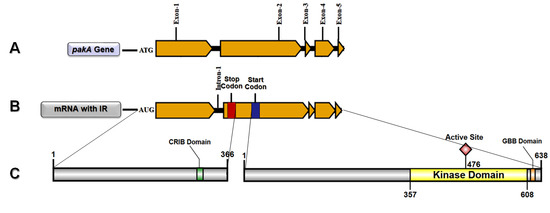
2.1. T. rubrum pakA/Ste20 Kinase Gene
2.2. Transcription Retention of Introns of the pakA Kinase Gene of T. rubrum
2.3. UDA Affects the Transcription Process of the pakA/Ste20 Gene in T. rubrum
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. RNA Extraction, cDNA Library Construction, and High-Throughput Sequencing
4.3. Data Analysis
4.4. RT-PCR and qRT-PCR Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMD | Amidation |
| CRIB | Cdc42/Rac interactive binding |
| FDR | False discovery rate |
| GBB | Gβ binding |
| IR | Intron retention |
| MAPK | Mitogen-activated protein kinase |
| MIC | Minimum inhibitory concentration |
| MYR | N-myristoylation |
| N-gly | N-glycosylation |
| PAK | p21 activated kinase |
| PTMs | Post-translational modifications |
| RNA-Seq | RNA-Sequencing |
| RT-PCR | Reverse transcription polymerase chain reaction |
| qRT-PCR | Quantitative reverse transcription-PCR |
| UDA | Undecanoic acid |
References
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. Microbiol. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Zeilinger, S.; Gruber, S.; Bansal, R.; Mukherjee, P.K. Secondary metabolism in Trichoderma–Chemistry meets genomics. Fungal Biol. Rev. 2016, 30, 74–90. [Google Scholar] [CrossRef]
- Kosti, I.; Mandel-Gutfreund, Y.; Glaser, F.; Horwitz, B.A. Comparative analysis of fungal protein kinases and associated domains. BMC Genomics 2010, 11, 133. [Google Scholar] [CrossRef] [PubMed]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Arana, D.M.; Nombela, C.; Alonso-Monge, R.; Pla, J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 2007, 15, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Manser, E.; Leung, T.; Salihuddin, H.; Zhao, Z.S.; Lim, L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 1994, 367, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; Andrianopoulos, A. Ste20-related kinases: Effectors of signaling and morphogenesis in fungi. Trends Microbiol. 2011, 19, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Rudel, T.; Bokoch, G.M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science 1997, 276, 1571–1574. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Davey, M.; Hsu, Y.C.; Kaplanek, P.; Tong, A.; Parsons, A.B.; Krogan, N.; Cagney, G.; Mai, D.; Greenblatt, J.; et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 2005, 120, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.H.; Johnson, D.A.; Traugh, J.A. Analysis of conformational changes during activation of protein kinase Pak2 by amide hydrogen/deuterium exchange. J. Biol. Chem. 2008, 283, 36397–363405. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.I.; Bhattacharya, A.; Wang, W.; Peltz, S.W. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 2001, 274, 15–25. [Google Scholar] [CrossRef]
- Vermout, S.; Tabart, J.; Baldo, A.; Mathy, A.; Losson, B.; Mignon, B. Pathogenesis of dermatophytosis. Mycopathologia 2008, 166, 267–275. [Google Scholar] [CrossRef] [PubMed]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Castro, O.; Movsichoff, F.; Parodi, A.J. Preferential transfer of the complete glycan is determined by the oligosaccharyltransferase complex and not by the catalytic subunit. Proc. Natl. Acad. Sci. USA 2006, 103, 14756–14760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lien, E.C.; Nagiec, M.J.; Dohlman, H.G. Proper protein glycosylation promotes mitogen-activated protein kinase signal fidelity. Biochemistry 2013, 52, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Sells, M.A.; Chernoff, J. Emerging from the Pak: The p21-activated protein kinase family. Trends Cell Biol. 1997, 7, 162–167. [Google Scholar] [CrossRef]
- Lin, M.; Grillitsch, K.; Daum, G.; Just, U.; Hofken, T. Modulation of sterol homeostasis by the Cdc42p effectors Cla4p and Ste20p in the yeast Saccharomyces cerevisiae. FEBS J. 2009, 276, 7253–7264. [Google Scholar] [CrossRef] [PubMed]
- Leberer, E.; Dignard, D.; Harcus, D.; Thomas, D.Y.; Whiteway, M. The Protein-Kinase Homolog Ste20p Is Required to Link the Yeast Pheromone Response G-Protein Beta-Gamma Subunits to Downstream Signaling Components. EMBO J. 1992, 11, 4815–4824. [Google Scholar] [CrossRef] [PubMed]
- Van Drogen, F.; O’Rourke, S.M.; Stucke, V.M.; Jaquenoud, M.; Neiman, A.M.; Peter, M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000, 10, 630–639. [Google Scholar] [CrossRef]
- Song, J.; Chen, Z.; Xu, P.; Gingras, R.; Ng, A.; Leberer, E.; Thomas, D.Y.; Ni, F. Molecular interactions of the Gbeta binding domain of the Ste20p/PAK family of protein kinases. An isolated but fully functional Gbeta binding domain from Ste20p is only partially folded as shown by heteronuclear NMR spectroscopy. J. Biol. Chem. 2001, 276, 41205–41212. [Google Scholar] [CrossRef] [PubMed]
- Merida, I.; Williamson, P.; Kuziel, W.A.; Greene, W.C.; Gaulton, G.N. The serine-rich cytoplasmic domain of the interleukin-2 receptor beta chain is essential for interleukin-2-dependent tyrosine protein kinase and phosphatidylinositol-3-kinase activation. J. Biol. Chem. 1993, 268, 6765–6770. [Google Scholar] [PubMed]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belvitch, P.; Adyshev, D.; Elangovan, V.R.; Brown, M.E.; Naureckas, C.; Rizzo, A.N.; Siegler, J.H.; Garcia, J.G.; Dudek, S.M. Proline-rich region of non-muscle myosin light chain kinase modulates kinase activity and endothelial cytoskeletal dynamics. Microvasc. Res. 2014, 95, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, L.; Hastwell, P.W.; Keenan, P.O.; Knight, A.W.; Billinton, N.; Walmsley, R.M. Genetic modification and variations in solvent increase the sensitivity of the yeast RAD54-GFP genotoxicity assay. Mutagenesis 2005, 20, 317–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audagnotto, M.; Dal Peraro, M. Protein post-translational modifications: In silico prediction tools and molecular modeling. Comput. Struct. Biotechnol. J. 2017, 15, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Hantschel, O.; Nagar, B.; Guettler, S.; Kretzschmar, J.; Dorey, K.; Kuriyan, J.; Superti-Furga, G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell 2003, 112, 845–857. [Google Scholar] [CrossRef]
- Gaffarogullari, E.C.; Masterson, L.R.; Metcalfe, E.E.; Traaseth, N.J.; Balatri, E.; Musa, M.M.; Mullen, D.; Distefano, M.D.; Veglia, G. A myristoyl/phosphoserine switch controls cAMP-dependent protein kinase association to membranes. J. Mol. Biol. 2011, 411, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Boutin, J.A. Myristoylation. Cell Signal 1997, 9, 15–35. [Google Scholar] [CrossRef]
- Patwardhan, P.; Resh, M.D. Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol. Cell Biol. 2010, 30, 4094–4107. [Google Scholar] [CrossRef] [PubMed]
- Eipper, B.A.; Stoffers, D.A.; Mains, R.E. The biosynthesis of neuropeptides: Peptide alpha-amidation. Annu. Rev. Neurosci. 1992, 15, 57–85. [Google Scholar] [CrossRef] [PubMed]
- Chufán, E.E.; De, M.; Eipper, B.A.; Mains, R.E.; Amzel, L.M. Amidation of bioactive peptides: The structure of the lyase domain of the amidating enzyme. Structure 2009, 17, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Niu, S.; Zheng, L.; Hu, L.; Huang, T.; Gu, L.; Feng, K.; Zhang, N.; Cai, Y.; Li, Y. Prediction of protein amidation sites by feature selection and analysis. Mol. Genet. Genomics 2013, 288, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Beenstock, J.; Mooshayef, N.; Engelberg, D. How Do Protein Kinases Take a Selfie (Autophosphorylate)? Trends Biochem. Sci. 2016, 41, 938–953. [Google Scholar] [CrossRef] [PubMed]
- Walter, B.N.; Huang, Z.; Jakobi, R.; Tuazon, P.T.; Alnemri, E.S.; Litwack, G.; Traugh, J.A. Cleavage and activation of p21-activated protein kinase gamma-PAK by CPP32 (caspase 3). Effects of autophosphorylation on activity. J. Biol. Chem. 1998, 273, 28733–28739. [Google Scholar] [CrossRef] [PubMed]
- Leal, J.; Squina, F.M.; Freitas, J.S.; Silva, E.M.; Ono, C.J.; Martinez-Rossi, N.M.; Rossi, A. A splice variant of the Neurospora crassa hex-1 transcript, which encodes the major protein of the Woronin body, is modulated by extracellular phosphate and pH changes. FEBS Lett. 2009, 583, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, G.L.; Oliveira, E.H.; Peres, N.T.A.; Cruz, A.H.; Martinez-Rossi, N.M.; Rossi, A. Transcription of Aspergillus nidulans pacC is modulated by alternative RNA splicing of palB. FEBS Lett. 2011, 585, 3442–3445. [Google Scholar] [CrossRef] [PubMed]
- Mendes, N.S.; Silva, P.M.; Silva-Rocha, R.; Martinez-Rossi, N.M.; Rossi, A. Pre-mRNA splicing is modulated by antifungal drugs in the filamentous fungus Neurospora crassa. FEBS Open Bio 2016, 6, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Li, G.; Yu, D.; Huang, W.; Cheng, C.; Liao, S.; Wu, Q.; Zhang, Y. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae. BMC Genomics 2017, 18, 130. [Google Scholar] [CrossRef] [PubMed]
- Mendes, N.S.; Bitencourt, T.A.; Sanches, P.R.; Silva-Rocha, R.; Martinez-Rossi, N.M.; Rossi, A. Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Sci. Rep. 2018, 8, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Juneau, K.; Nislow, C.; Davis, R.W. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 2009, 183, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013, 497, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.; Meng, X.; Zhao, Z.; Kang, D.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.; Wells, R.; Rechsteiner, M. Amino acid sequences common to rapidly degraded proteins: The PEST hypothesis. Science 1986, 234, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Leberer, E.; Harcus, D.; Broadbent, I.D.; Clark, K.L.; Dignard, D.; Ziegelbauer, K.; Schmidt, A.; Gow, N.A.R.; Brown, A.J.P.; Thomas, D.Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 1996, 93, 13217–13222. [Google Scholar] [CrossRef] [PubMed]
- Leberer, E.; Ziegelbauer, K.; Schmidt, A.; Harcus, D.; Dignard, D.; Ash, J.; Johnson, L.; Thomas, D.Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr. Biol. 1997, 7, 539–546. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2012, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Bourgon, R.; Gentleman, R.; Huber, W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. USA 2010, 107, 9546–9551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s CT difference” formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Jacob, T.R.; Peres, N.T.; Persinoti, G.F.; Silva, L.G.; Mazucato, M.; Rossi, A.; Martinez-Rossi, N.M. rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 2012, 50, 368–377. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, E.V.; Bortolossi, J.C.; Sanches, P.R.; Mendes, N.S.; Martinez-Rossi, N.M.; Rossi, A. STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum. Int. J. Mol. Sci. 2018, 19, 3654. https://doi.org/10.3390/ijms19113654
Gomes EV, Bortolossi JC, Sanches PR, Mendes NS, Martinez-Rossi NM, Rossi A. STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum. International Journal of Molecular Sciences. 2018; 19(11):3654. https://doi.org/10.3390/ijms19113654
Chicago/Turabian StyleGomes, Eriston V., Julio C. Bortolossi, Pablo R. Sanches, Niege S. Mendes, Nilce M. Martinez-Rossi, and Antonio Rossi. 2018. "STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum" International Journal of Molecular Sciences 19, no. 11: 3654. https://doi.org/10.3390/ijms19113654
APA StyleGomes, E. V., Bortolossi, J. C., Sanches, P. R., Mendes, N. S., Martinez-Rossi, N. M., & Rossi, A. (2018). STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum. International Journal of Molecular Sciences, 19(11), 3654. https://doi.org/10.3390/ijms19113654