Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications
Abstract
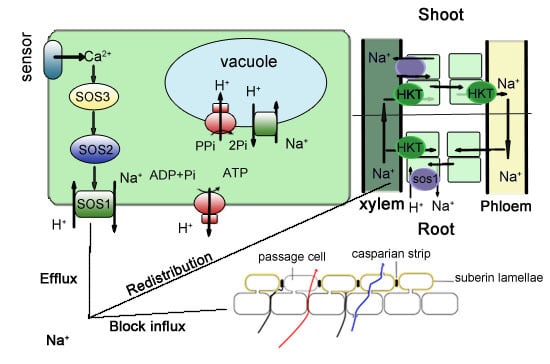
:1. Introduction
2. Control of Na+ Uptake
2.1. Blocking Na+ Influx into the Root
2.2. Enhancing the Na+ Efflux in the Root
3. Control of Na+ Distribution
3.1. Reducing Na+ Transport to Shoots and Distribution to Roots and Root–Stem Junctions
3.2. Na+ Distribution to Specific Parts of Shoots
3.3. Na+ Recirculation in the Phloem
4. Using Improved Salt Tolerance to Improve Crops
4.1. Exploiting the Key Genes Responsible for Forming Apoplastic Barrier (Especially the Casparian Strip)
4.2. Increasing the Activities of the SOS Pathway and the H+-ATPase
4.3. Increasing the Activities of HKT-Like Transporters
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Feng, Z.T.; Deng, Y.Q.; Zhang, S.C.; Liang, X.; Yuan, F.; Hao, J.L.; Zhang, J.C.; Sun, S.F.; Wang, B.S. K+ accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 2015, 238, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.H.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.; Wang, M.; Yuan, F.; Sui, Na.; Song, J.; Wang, B. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2014, 86, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Smajgl, A.; Toan, T.Q.; Nhan, D.K.; Ward, J.; Trung, N.H.; Tri, L.Q.; Tri, V.P.D.; Vu, P.T. Responding to rising sea levels in the Mekong Delta. Nat. Clim. Change 2015, 5, 167–174. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.P.; Yuan, F.; Yang, Z.; Wang, B.S.; Chen, M. Transcriptome profiling of genes involved in photosynthesis in Elaeagnus angustifolia L. under salt stress. Photosynthetica 2018, 56, 998–1009. [Google Scholar] [CrossRef]
- Shabala, S.; Bose, J.; Hedrich, R. Salt bladders: Do they matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Wei, X.; Zhao, X.; Wang, B.; Sui, N. Transcription profiles of genes related to hormonal regulations under salt stress in sweet sorghum. Plant Mol. Biol. Rep. 2017, 35, 586–599. [Google Scholar] [CrossRef]
- Flowers, T.J. Halophytes. In Ion Transport in Plant Cell and Tissues; Baker, D.A., Hall, J.L., Eds.; American Elsevier: New York, NY, USA, 1975; pp. 309–334. [Google Scholar]
- Jennings, D.H. The effect of sodium chloride on higher plants. Diol. Rev. 1976, 51, 453–486. [Google Scholar] [CrossRef]
- Richards, L.A. Diagnosis and improvement of saline and alkali soils. Soil Sci. 1954, 78, 154. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, Q.; Deng, Y.; Sun, S.; Zhang, J.; Wang, B. Study on pathway and characteristics of ion secretion of salt glands of Limonium bicolor. Acta Physiol. Plant 2014, 10, 2729–2741. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot-London 2015, 115, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B.S. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct. Plant Biol. 2018, 45, 350–361. [Google Scholar] [CrossRef]
- Breckle, S.W. How do halophytes overcome salinity. In Biology of Salt Tolerant Plants; Ajmal, K.M., Irwin, A.U., Eds.; Book Crafters: Parker, CO, USA, 1995; pp. 199–213. [Google Scholar]
- Deng, Y.; Feng, Z.; Yuan, F.; Guo, J.; Suo, S.; Wang, B. Identification and functional analysis of the autofluorescent substance in Limonium bicolor salt glands. Plant Physiol. Bioch. 2015, 97, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lyu, M.A.; Leng, B.Y.; Zheng, G.Y.; Feng, Z.T.; Li, P.H.; Zhu, X.G.; Wang, B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015, 38, 1637–1657. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lyu, M.J.; Leng, B.Y.; Zhu, X.G.; Wang, B.S. The transcriptome of NaCl-treated Limoniumbicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. Rep. 2016, 91, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Leng, B.; Wang, B. Progress in studying salt secretion from the salt glands in recretohalophytes: How do plants secrete salt? Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.S.; Lutz, A.; Rupasinghe, T.; Bazihizina, N.; Bohm, J.; Alharbi, S.; Hedrich, R.; Shabala, S. Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 2017, 40, 1900–1915. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Song, J.; Wang, B.S. NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiol. Plant 2010, 32, 27–36. [Google Scholar] [CrossRef]
- Cheng, S.; Yang, Z.; Wang, M.; Song, J.; Sui, N.; Fan, H. Salinity improves chilling resistance in Suaeda salsa. Acta Physiol. Plant 2014, 36, 1823–1830. [Google Scholar] [CrossRef]
- Sui, N. Photoinhibition of Suaeda salsa to chilling stress is related to energy dissipation and water-water cycle. Photosynthetica 2015, 53, 207–212. [Google Scholar] [CrossRef]
- Chen, T.; Yuan, F.; Song, J.; Wang, B. Nitric oxide participates in waterlogging tolerance through enhanced adventitious root formation in the euhalophyte Suaeda salsa. Funct. Plant Biol. 2016, 43, 244–253. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, R.; Sui, N.; Shi, W.; Wang, L.; Tian, C.; Song, J. Changes in endogenous hormones and seed-coat phenolics during seed storage of two Suaeda salsa populations. Aust. J. Bot. 2016, 64, 325–332. [Google Scholar] [CrossRef]
- Matsushita, N.; Matoh, T. Characterization of Na+ exclusion mechanisms of salt-tolerant reed plants in comparison with salt-sensitive rice plants. Physiol. Plant 1991, 83, 170–176. [Google Scholar] [CrossRef]
- Flowers, T.J.; Galal, H.K.; Bromham, L. Evolution of halophytes: Multiple origins of salt tolerance in land plants. Funct. Plant Biol. 2010, 37, 604–612. [Google Scholar] [CrossRef]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Al-Azzawi, M.; Aronson, J.; Flowers, T.J. eHALOPH a database of salt-tolerant plants: Helping put halophytes to work. Plant Cell Physiol. 2016, 57, e10. [Google Scholar] [CrossRef] [PubMed]
- Levitt, J. Responses of Plant to Environmental Stress Chilling, Freezing, and High Temperature Stresses, 2nd ed.; Academic Press: New York, NY, USA, 1980; p. 497. [Google Scholar]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Husain, S.; Von Caemmerer, S.; Munns, R. Control of salt transport from roots to shoots of wheat in saline soil. Funct. Plant Biol. 2004, 31, 1115–1126. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheeseman, J.M. Pump-leak sodium fluxes in low salt corn roots. J. Membrane Biol. 1982, 70, 157–164. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot-London 2003, 91, 503–527. [Google Scholar] [CrossRef]
- Ochiai, K.; Matoh, T. Characterization of the Na+ delivery from roots to shoots in rice under saline stress: Excessive salt enhances apoplastic transport in rice plants. Soil Sci. Plant Nutr. 2002, 48, 371–378. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Ranathunge, K.; Nayak, S.; Schreiber, L.; Mathew, M. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 4215–4228. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, L.; Zeier, J. Apoplastic barriers in roots: Chemical composition of endodermal and hypodermal cell walls. J. Exp. Bot. 1999, 50, 1267–1280. [Google Scholar] [CrossRef]
- Yeo, A.; Yeo, M.; Flowers, T. The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 1987, 38, 1141–1153. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Barberon, M.; Geldner, N. Radial transport of nutrients: The plant root as a polarized epithelium. J. Plant Physiol. 2014, 66, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Ranathunge, K.; Franke, R.; Prakash, H.; Schreiber, L.; Mathew, M. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 2009, 230, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; yothi-Prakash, P.A.; Qin, L.; He, J.; Lin, Q.; Loh, C.S.; Kumar, P.P. Role of root hydrophobic barriers in salt exclusion of a mangrove plant Avicennia officinalis. Plant Cell Environ. 2014, 37, 1656–1671. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Francini, A.; Minnocci, A.; Sebastiani, L. Salt stress modifies apoplastic barriers in olive (Olea europaea L.): A comparison between a salt-tolerant and a salt-sensitive cultivar. Sci. Hortic-Amsterdam 2015, 192, 38–46. [Google Scholar] [CrossRef]
- Shen, J.; Xu, G.; Zheng, H.Q. Apoplastic barrier development and water transport in Zea mays seedling roots under salt and osmotic stresses. Protoplasma 2015, 252, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Tylová, E.; Pecková, E.; Blascheová, Z.; Soukup, A. Casparian bands and suberin lamellae in exodermis of lateral roots: An important trait of roots system response to abiotic stress factors. Ann. Bot-London 2017, 120, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin biosynthesis and structure. J. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Ranathunge, K.; Schreiber, L.; Franke, R. Suberin research in the genomics era—New interest for an old polymer. Plant Sci. 2011, 180, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Franke, R.; Schreiber, L. Suberin-a biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 2007, 10, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Jung, S.J.; Go, Y.S.; Kim, H.U.; Kim, J.K.; Cho, H.J.; Park, O.K.; Suh, M.C. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009, 60, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, I.; Bronner, R.; Wang, Y.; Compagnon, V.; Michler, P.; Schreiber, L.; Salaün, J.P.; Durst, F.; Pinot, F. CYP94A1, a plant cytochrome P450-catalyzing fatty acid ω-hydroxylase, is selectively induced by chemical stress in Vicia sativa seedlings. Planta 2005, 221, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, I.; Tijet, N.; Adas, F.; Philipps, G.; Salaün, J.P.; Durst, F. CYP86A1 from Arabidopsis thaliana Encodes a Cytochrome P450-Dependent Fatty Acid Omega-Hydroxylase. BBRC 1998, 243, 688–693. [Google Scholar] [PubMed]
- Compagnon, V.; Diehl, P.; Benveniste, I.; Meyer, D.; Schaller, H.; Schreiber, L.; Franke, R.; Pinot, F. CYP86B1 is required for very long chain ω-hydroxyacid and α, ω-dicarboxylic acid synthesis in root and seed suberin polyester. J. Plant Physiol. 2009, 150, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef] [PubMed]
- Bouquin, R.L.; Pinot, F.; Benveniste, I.; Salaün, J.P.; Durst, F. Cloning and functional characterization of CYP94A2, a medium chain fatty acid hydroxylase from Vicia sativa. Biochem. Biophres. Co. 1999, 261, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Hosmani, P.S.; Rus, A.; Lahner, B.; Borevitz, J.O.; Muthukumar, B.; Mickelbart, M.V.; Schreiber, L.; Franke, R.B.; Salt, D.E. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009, 5, 188–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosmani, P.S.; Kamiya, T.; Danku, J.; Naseer, S.; Geldner, N.; Guerinot, M.L.; Salt, D.E. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. USA 2013, 110, 14498–14503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhu, J.K. A calcium sensor homolog required for plant salt tolerance. Science 1998, 280, 1943–1945. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ishitani, M.; Kim, C.S.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Morsomme, P.; Boutry, M. The plant plasma membrane H+ -ATPase: Structure, function and regulation. BBA-Biomembranes 2000, 1465, 1–16. [Google Scholar] [CrossRef]
- Palmgren, M.G. Plant plasma membrane H+-ATPases: Power-houses for nutrient uptake. Plant Mol. Biol. 2001, 52, 817–845. [Google Scholar]
- Yang, Y.; Qin, Y.; Xie, C.; Zhao, F.; Zhao, J.; Liu, D.; Chen, S.; Fuglsang, A.T.; Palmgren, M.G.; Schumaker, K.S.; et al. The Arabidopsis Chaperone J3 Regulates the Plasma Membrane H+-ATPase through Interaction with the PKS5 Kinase. Plant Cell 2010, 22, 1313–1332. [Google Scholar] [CrossRef] [PubMed]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Han, N.; Ding, T.; Zhou, F.; Wang, B. SsHKT1;1 is a potassium transporter of the C 3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 2014, 41, 790–802. [Google Scholar] [CrossRef]
- Jaime-Pérez, N.; Pineda, B.; García-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olías, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A Retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2017, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, S.; Horie, T.; Hauser, F.; Deinlein, U.; Schroeder, J.I.; Uozumi, N. HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotech. 2015, 32, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The OsHKT1; 1 transporter is involved in salt tolerance and regulated by an MYB-type transcription factor. J. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y. OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon slat stress. BMC Plant Biol. 2016, 16, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1; 5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A Magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice. J. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Xu, B.; Krishnan, M.; Lightfoot, D.J.; Athman, A.; Jacobs, A.K.; Watson-Haigh, N.S.; Plett, D.; Munns, R.; Tester, M.; et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 2014, 80, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yuan, F.; Wang, B.S.; Chen, M. Response of energy plant hybrid pennisetum to NaCl stress and its salinity threshold. Chinese J. Plant Ecol. 2012, 36, 572–577. [Google Scholar] [CrossRef]
- Davenport, R.; James, R.A.; Zakrisson-Plogander, A.; Tester, M.; Munns, R. Control of sodium transport in durum wheat. Plant Physiol. 2005, 137, 807–818. [Google Scholar] [CrossRef] [PubMed]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterisation of two genes for Na+ exclusion in durum wheat: Nax1 and Nax2. Plant Physiol. 2006, 142, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.R. HKT1; 5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. J. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Gilliham, M.; Able, J.A.; Roy, S.J. Translating knowledge in abiotic stress tolerance to breeding programs. Plant. J. 2016, 90, 898–917. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 2010, 28, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Lee, B.; Wu, S.J.; Zhu, J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Xiaocen, W.; Baoshan, W. Sorghum bicolor (L.) Moench, is a variation of traditional grain sorghum; Shandong Normal University: Jinan, China, 2017. [Google Scholar]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Yang, Z.; Liu, J.; Zhu, T.; Wei, X.; Fan, H.; Wang, B. Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications. Int. J. Mol. Sci. 2018, 19, 3668. https://doi.org/10.3390/ijms19113668
Chen M, Yang Z, Liu J, Zhu T, Wei X, Fan H, Wang B. Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications. International Journal of Molecular Sciences. 2018; 19(11):3668. https://doi.org/10.3390/ijms19113668
Chicago/Turabian StyleChen, Min, Zhen Yang, Jing Liu, Tingting Zhu, Xiaocen Wei, Hai Fan, and Baoshan Wang. 2018. "Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications" International Journal of Molecular Sciences 19, no. 11: 3668. https://doi.org/10.3390/ijms19113668
APA StyleChen, M., Yang, Z., Liu, J., Zhu, T., Wei, X., Fan, H., & Wang, B. (2018). Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications. International Journal of Molecular Sciences, 19(11), 3668. https://doi.org/10.3390/ijms19113668