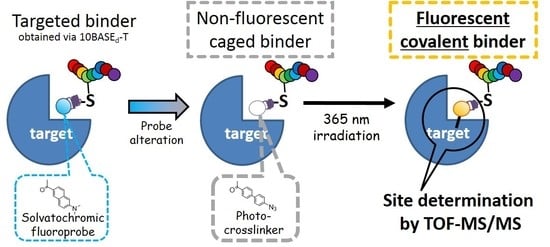
A Cysteine-Reactive Small Photo-Crosslinker Possessing Caged-Fluorescence Properties: Binding-Site Determination of a Combinatorially-Selected Peptide by Fluorescence Imaging/Tandem Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of Cysteine-Reactive Small Photo-Crosslinkers and their Attachment to the Combinatorially-Selected Peptide
2.2. Targeted Covalent Conjugation of Caged Binder with GST
2.3. Molecular Docking Simulation for Rationalizing Cross-Linking Specificity
3. Materials and Methods
3.1. General
3.2. Photo-Crosslinking of the Caged Binder and GST
3.3. In-Gel Trypsinization of Crosslinked GST and Mass Spectrometric Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| MALDI-TOF-MS/MS | Matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry |
| Prodan | 6-Propionyl-2-dimethylaminonaphthalene |
| UV | Ultraviolet |
| DMSO | Dimethyl sulfoxide |
| D-PBS | Dulbecco’s phosphate-buffered saline |
| PDB | Protein Data Bank |
References
- Pazos, E.; Vazquez, O.; Mascarenas, J.L.; Vazquez, M.E. Peptide-based fluorescent biosensors. Chem. Soc. Rev. 2009, 38, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Choulier, L.; Enander, K. Environmentally Sensitive Fluorescent Sensors Based on Synthetic Peptides. Sensors 2010, 10, 3126–3144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zhu, L.P.; Hirano, Y.; Karirninavargani, M.; Tada, S.; Zhang, G.X.; Uzawa, T.; Zhang, D.Q.; Hirose, T.; Taiji, M.; et al. Fluorogenic Enhancement of an in Vitro-Selected Peptide Ligand by Replacement of a Fluorescent Group. Anal. Chem. 2016, 88, 7991–7997. [Google Scholar] [CrossRef] [PubMed]
- Taki, M.; Inoue, H.; Mochizuki, K.; Yang, J.; Ito, Y. Selection of Color-Changing and Intensity-Increasing Fluorogenic Probe as Protein-Specific Indicator Obtained via the 10BASE(d)-T. Anal. Chem. 2016, 88, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Uzawa, T.; Tochio, N.; Hamatsu, J.; Hirano, Y.; Tada, S.; Saneyoshi, H.; Kigawa, T.; Hayashi, N.; Ito, Y.; et al. A fluorogenic peptide probe developed by in vitro selection using tRNA carrying a fluorogenic amino acid. Chem. Commun. 2014, 50, 2962–2964. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chung, N.N.; Lemieux, C.; Zelent, B.; Vanderkooi, J.M.; Gryczynski, I.; Wilkes, B.C.; Schiller, P.W. [Aladan3]TIPP: A fluorescent delta-opioid antagonist with high delta-receptor binding affinity and delta selectivity. Biopolymers 2005, 80, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Svensson, R.; Greno, C.; Johansson, A.S.; Mannervik, B.; Morgenstern, R. Synthesis and characterization of 6-chloroacetyl-2-dimethylaminonaphthalene as a fluorogenic substrate and a mechanistic probe for glutathione transferases. Anal. Biochem. 2002, 311, 171–178. [Google Scholar] [CrossRef]
- Gonzalez-Vera, J.A.; Morris, M.C. Fluorescent Reporters and Biosensors for Probing the Dynamic Behavior of Protein Kinases. Proteomes 2015, 3, 369–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeno, T.; Uzawa, T.; Kono, I.; Okano, K.; Iino, T.; Fukita, K.; Oshikawa, Y.; Ogawa, T.; Iwata, O.; Ito, T.; et al. Targeted delivery of fluorogenic peptide aptamers into live microalgae by femtosecond laser photoporation at single- cell resolution. Sci. Rep. 2018, 8, 8271. [Google Scholar] [CrossRef] [PubMed]
- Loving, G.; Imperiali, B. Thiol-Reactive Derivatives of the Solvatochromic 4-N,N-Dimethylamino-1,8-naphthalimide Fluorophore: A Highly Sensitive Toolset for the Detection of Biomolecular Interactions. Bioconj. Chem. 2009, 20, 2133–2141. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, S.; Tabuchi, Y.; Ito, Y.; Taki, M. Combinatorially Screened Peptide as Targeted Covalent Binder: Alteration of Bait-Conjugated Peptide to Reactive Modifier. Bioconj. Chem. 2018, 29, 1866–1871. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, R.; Renard, B.Y. Docking small peptides remains a great challenge: an assessment using AutoDock Vina. Brief. Bioinform. 2015, 16, 1045–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziarek, J.J.; Peterson, F.C.; Lytle, B.L.; Volkman, B.F. Binding Site Identification and Structure Determination of Protein-Ligand Complexes by Nmr: A Semiautomated Approach. Method Enzymol. 2011, 493, 241–275. [Google Scholar] [CrossRef]
- Artigues, A.; Nadeau, O.W.; Rimmer, M.A.; Villar, M.T.; Du, X.X.; Fenton, A.W.; Carlson, G.M. Protein Structural Analysis via Mass Spectrometry-Based Proteomics. In Modern Proteomic—Sample Preparation, Analysis and Practical Applications; Springer: Cham, Switzerland, 2016; Volume 919, pp. 397–431. [Google Scholar] [CrossRef]
- Serpa, J.J.; Parker, C.E.; Petrotchenko, E.V.; Han, J.; Pan, J.X.; Borchers, C.H. Mass spectrometry-based structural proteomics. Eur. J. Mass Spectrom. 2012, 18, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.W.; Radford, S.E.; Ashcroft, A.E.; Wilson, A.J. Analysis of amyloid nanostructures using photo-cross-linking: In situ comparison of three widely used photo-cross-linkers. ACS Chem. Biol. 2014, 9, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.W.; Wilson, A.J. Photo-induced covalent cross-linking for the analysis of biomolecular interactions. Chem. Soc. Rev. 2013, 42, 3289–3301. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.-S.; Chen, B.; Wu, Y.-Y.; Long, Q.-S.; Zhao, Y.-L.; Wang, P.-Y.; Yang, S. Current advances of carbene-mediated photoaffinity labeling in medicinal chemistry. RSC Adv. 2018, 8, 29428–29454. [Google Scholar] [CrossRef]
- Murale, D.P.; Hong, S.C.; Haque, M.M.; Lee, J.S. Photo-affinity labeling (PAL) in chemical proteomics: A handy tool to investigate protein-protein interactions (PPIs). Proteome Sci. 2016, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Xin, B.T.; van Tol, B.D.M.; Ovaa, H.; Geurink, P.P. Native chemical ligation at methionine bioisostere norleucine allows for N-terminal chemical protein ligation. Org. Biomol. Chem. 2018, 16, 6306–6315. [Google Scholar] [CrossRef] [PubMed]
- Ota, E.; Usui, K.; Oonuma, K.; Koshino, H.; Nishiyama, S.; Hirai, G.; Sodeoka, M. Thienyl-Substituted alpha-Ketoamide: A Less Hydrophobic Reactive Group for Photo-Affinity Labeling. ACS Chem. Biol. 2018, 13, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Asanuma, M.; Ishikawa, M.; Hashimoto, Y.; Dodo, K.; Sodeoka, M.; Yamaguchi, T. Specific fluorescence labeling of target proteins by using a ligand-4-azidophthalimide conjugate. Chem. Commun. 2017, 53, 8751–8754. [Google Scholar] [CrossRef] [PubMed]
- Tomohiro, T.; Morimoto, S.; Shima, T.; Chiba, J.; Hatanaka, Y. An Isotope-Coded Fluorogenic Cross-Linker for High-Performance Target Identification Based on Photoaffinity Labeling. Angew. Chem. Int. Ed. 2014, 53, 13502–13505. [Google Scholar] [CrossRef] [PubMed]
- Soethoudt, M.; Stolze, S.C.; Westphal, M.V.; van Stralen, L.; Martella, A.; van Rooden, E.J.; Guba, W.; Varga, Z.V.; Deng, H.; van Kasteren, S.I.; et al. Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB2 Receptor Expression and Ligand Engagement in Human Cells. J. Am. Chem. Soc. 2018, 140, 6067–6075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapinsky, D.J. Tandem photoaffinity labeling-bioorthogonal conjugation in medicinal chemistry. Bioorg. Med. Chem. 2012, 20, 6237–6247. [Google Scholar] [CrossRef] [PubMed]
- Kepiro, M.; Varkuti, B.H.; Rauscher, A.A.; Kellermayer, M.S.Z.; Varga, M.; Malnasi-Csizmadia, A. Molecular tattoo: Subcellular confinement of drug effects. Chem. Biol. 2015, 22, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Lord, S.J.; Lee, H.L.; Samuel, R.; Weber, R.; Liu, N.; Conley, N.R.; Thompson, M.A.; Twieg, R.J.; Moerner, W.E. Azido push-pull fluorogens photoactivate to produce bright fluorescent labels. J. Phys. Chem. B 2010, 114, 14157–14167. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Drummen, G.P.C.; Konishi, G. Recent advances in twisted intramolecular charge transfer (TICT) fluorescence and related phenomena in materials chemistry. J. Mater. Chem. C 2016, 4, 2731–2743. [Google Scholar] [CrossRef] [Green Version]
- Anzalone, A.V.; Chen, Z.; Cornish, V.W. Synthesis of photoactivatable azido-acyl caged oxazine fluorophores for live-cell imaging. Chem. Commun. 2016, 52, 9442–9445. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Collins, I. Photoaffinity labeling in target- and binding-site identification. Future Med. Chem. 2015, 7, 159–183. [Google Scholar] [CrossRef] [PubMed]
- Brase, S.; Gil, C.; Knepper, K.; Zimmermann, V. Organic azides: An exploding diversity of a unique class of compounds. Angew. Chem. Int. Ed. Engl. 2005, 44, 5188–5240. [Google Scholar] [CrossRef] [PubMed]
- Fukunishi, Y.; Mikami, Y.; Nakamura, H. Similarities among receptor pockets and among compounds: Analysis and application to in silico ligand screening. J. Mol. Graph. Model. 2005, 24, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Fukunishi, Y.; Mikami, Y.; Nakamura, H. The filling potential method: A method for estimating the free energy surface for protein-ligand docking. J. Phys. Chem. B 2003, 107, 13201–13210. [Google Scholar] [CrossRef]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yatabe, K.; Hisada, M.; Tabuchi, Y.; Taki, M. A Cysteine-Reactive Small Photo-Crosslinker Possessing Caged-Fluorescence Properties: Binding-Site Determination of a Combinatorially-Selected Peptide by Fluorescence Imaging/Tandem Mass Spectrometry. Int. J. Mol. Sci. 2018, 19, 3682. https://doi.org/10.3390/ijms19113682
Yatabe K, Hisada M, Tabuchi Y, Taki M. A Cysteine-Reactive Small Photo-Crosslinker Possessing Caged-Fluorescence Properties: Binding-Site Determination of a Combinatorially-Selected Peptide by Fluorescence Imaging/Tandem Mass Spectrometry. International Journal of Molecular Sciences. 2018; 19(11):3682. https://doi.org/10.3390/ijms19113682
Chicago/Turabian StyleYatabe, Kazuki, Masaru Hisada, Yudai Tabuchi, and Masumi Taki. 2018. "A Cysteine-Reactive Small Photo-Crosslinker Possessing Caged-Fluorescence Properties: Binding-Site Determination of a Combinatorially-Selected Peptide by Fluorescence Imaging/Tandem Mass Spectrometry" International Journal of Molecular Sciences 19, no. 11: 3682. https://doi.org/10.3390/ijms19113682
APA StyleYatabe, K., Hisada, M., Tabuchi, Y., & Taki, M. (2018). A Cysteine-Reactive Small Photo-Crosslinker Possessing Caged-Fluorescence Properties: Binding-Site Determination of a Combinatorially-Selected Peptide by Fluorescence Imaging/Tandem Mass Spectrometry. International Journal of Molecular Sciences, 19(11), 3682. https://doi.org/10.3390/ijms19113682