Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation, Characterization, and Identification of Bacterial Strains
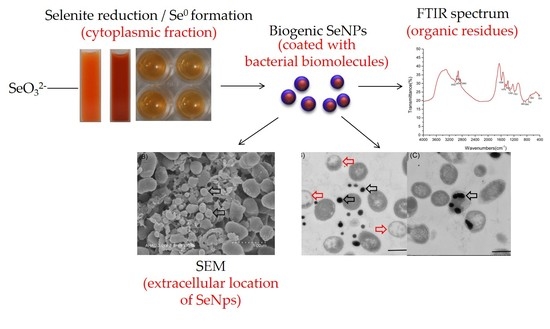
2.2. Bioconversion of SeO32− And Elemental Selenium Formation
2.3. Localization and Characterization of Selenium Nanoparticles
2.4. Characterization of SeNPs Produced by P. mirabilis YC801
2.4.1. Dynamic Light Scattering (DLS) Analyses and SEM- (Energy Dispersive X-ray) Analysis (EDX)
2.4.2. Fourier-Transform Infrared Spectroscopy (FTIR) Analysis
2.5. Biocatalytic Selenite Reduction Activity Assays
2.6. Real-Time PCR(qPCR) Analysis
3. Materials and Methods
3.1. Chemicals and Culture Medium
3.2. Bacterial Strain Isolation and Identification
3.3. Assessment of Sensitivity towards Selenite
3.4. Quantification of Selenite Reduction and Formation of Se0
3.4.1. Dynamics of Microbial Growth after Exposure to SeO32−
3.4.2. Assessment of SeO32− Bioreduction Efficiency
3.4.3. Se0 Content Analysis
3.5. Localization of SeNPs
3.5.1. TEM Analysis
3.5.2. SEM Analysis
3.6. Preparation and Characterization of SeNPs
3.6.1. Preparation of Biogenic SeNPs
3.6.2. DLS Analysis
3.6.3. SEM and EDX Analyses
3.6.4. FTIR Measurement
3.7. Detection of the Localization of Selenite Reduction Activity
3.7.1. Protein Extraction
3.7.2. EPS Extraction
3.7.3. Supernatant Preparation
3.7.4. Selenite Reduction Assay on Different Subcellular Fractions
3.8. qPCR
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SeNPs | Se nanoparticles |
| Se0 | Elemental Selenium |
| TEM | Transmission electron microscopy |
| SEM | Scanning electron microscopy |
| EDX | Energy dispersive X-ray analysis |
| MIC | Minimum inhibitory concentration |
| NADH | Nicotinamide adenine dinucleotide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| qPCR | Real-time quantitative PCR |
References
- Mishra, R.R.; Prajapati, S.; Das, J.; Dangar, T.K.; Das, N.; Thatoi, H. Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere 2011, 84, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Błażejak, S.; Bzducha-Wróbel, A.; Kot, A.M. Effect of selenium on lipid and amino acid metabolism in yeast cells. Biol. Trace Element Res. 2018. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, N.; Li, D.; He, S.; Chen, Y.; Bai, Y.; Zhou, M.; He, J.; Wang, C. Effects of Selenium on the Growth and Fermentation Properties of Se-Enriched Bacillus Subtilis J-2. J. Food Biochem. 2016, 40, 31–38. [Google Scholar] [CrossRef]
- Butler, C.S.; Debieux, C.M.; Dridge, E.J.; Splatt, P.; Wright, M. Biomineralization of selenium by the selenate-respiring bacterium Thauera selenatis. Biochem. Soc. Trans. 2012, 40, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Gojkovic, Z.; Garbayo, I.; Ariza, J.L.G.; Márová, I.; Vílchez, C. Selenium bioaccumulation and toxicity in cultures of green microalgae. Algal Res. 2015, 7, 106–116. [Google Scholar] [CrossRef]
- Han, D.; Xiong, S.; Tu, S.; Liu, J.; Chen, C. Interactive effects of selenium and arsenic on growth, antioxidant system, arsenic and selenium species of Nicotiana tabacum L. Environ. Exp. Bot. 2015, 117, 12–19. [Google Scholar] [CrossRef]
- Lee, K.H.; Jeong, D. Bimodal actions of selenium essential for antioxidant and toxic pro-oxidant activities: The selenium paradox (Review). Mol. Med. Rep. 2012, 5, 299–304. [Google Scholar] [PubMed]
- Bajaj, M.; Schmidt, S.; Winter, J. Formation of Se(0) Nanoparticles by Duganella sp. and Agrobacterium sp. isolated from Se-laden soil of North-East Punjab, India. Microb. Cell Fact. 2012, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Nancharaiah, Y.V.; Lens, P.N.L. Selenium biomineralization for biotechnological applications. Trends Biotechnol. 2015, 33, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Gallegos, M.; Doig, L.E.; Tse, J.J.; Pickering, I.J.; Liber, K. Bioavailability, Toxicity and Biotransformation of Selenium in Midge (Chironomus dilutus) Larvae Exposed via Water or Diet to Elemental Selenium Particles, Selenite, or Selenized Algae. Environ. Sci. Technol. 2013, 47, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef] [PubMed]
- Rovira, M.; Giménez, J.; Martínez, M.; Martínez-Lladó, X.; de Pablo, J.; Martí, V.; Duro, L. Sorption of selenium(IV) and selenium(VI) onto natural iron oxides: Goethite and hematite. J. Hazard. Mater. 2008, 150, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fu, F.; Zhang, L.; Tang, B. Insight into efficient co-removal of Se(IV) and Cr(VI) by magnetic mesoporous carbon microspheres: Performance and mechanism. Chem. Eng. J. 2018, 346, 590–599. [Google Scholar] [CrossRef]
- Zhang, M.; Reardon, E.J. Removal of B, Cr, Mo, and Se from wastewater by incorporation into hydrocalumite and ettringite. Environ. Sci. Technol. 2003, 37, 2947–2952. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Li, X.; Cheng, Y.; Xiao, X.; Lu, Z.; Wang, Y.; Wang, F. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 2017, 7, 3239. [Google Scholar] [CrossRef] [PubMed]
- Avendaño, R.; Chaves, N.; Fuentes, P.; Sánchez, E.; Jiménez, J.I.; Chavarría, M. Production of selenium nanoparticles in Pseudomonas putida KT2440. Sci. Rep. 2016, 6, 37155. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Biosynthesis of selenium nanoparticles by Azoarcus sp. CIB. Microb. Cell Fact. 2016, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Bernardi, P.; Butler, C.S.; Vallini, G. Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeITE01 as a consequence of selenite reduction under aerobic conditions. Microb. Cell Fact. 2014, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Błażejak, S.; Gientka, I.; Bzducha-Wróbel, A. Accumulation and metabolism of selenium by yeast cells. Appl. Microbiol. Biotechnol. 2015, 99, 5373–5382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, C.A.; Ridley, H.; Condie, K.L.; Leaver, J.T.; Richardson, D.J.; Butler, C.S. Selenate reduction by Enterobacter cloacae SLD1a-1 is catalysed by a molybdenum-dependent membrane-bound enzyme that is distinct from the membrane-bound nitrate reductase. FEMS Microbiol. Lett. 2003, 228, 273–279. [Google Scholar] [CrossRef]
- Jiménez-lamana, J.; Abad-álvaro, I.; Katarzyna, B.; Laborda, F.; Szpunar, J.; Lobinski, R. Detection and characterization of biogenic selenium nanoparticles in selenium-rich yeast by single particle ICPMS. J. Anal. At. Spectrom. 2018, 33, 452. [Google Scholar] [CrossRef]
- Eswayah, A.S.; Smith, T.J.; Gardiner, P.H. Microbial Transformations of Selenium Species of Relevance to Bioremediation. Appl. Environ. Microbiol. 2016, 82, 4848–4859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charya, L.S. Selenium pollution in the marine environment and marine bacteria in selenium bioremediation. In Marine Pollution and Microbial Remediation; Naik, M., Dubey, S., Eds.; Springer: Singapore, 2017; pp. 223–237. [Google Scholar]
- Lenz, M.; Lens, P.N. The essential toxin: The changing perception of selenium in environmental sciences. Sci. Total Environ. 2009, 407, 3620–3633. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kim, Y.J.; Zhang, D.; Yang, D.C. Biological Synthesis of Nanoparticles from Plants and Microorganisms. Trends Biotechnol. 2016, 34, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, D.; Pan, X.; Lee, D.J.; Al-Misned, F.A.; Mortuza, M.G.; Gadd, G.M. Aerobic and anaerobic biosynthesis of nano-selenium for remediation of mercury contaminated soil. Chemosphere 2016, 170, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.A.; Webster, T.J. Selenium nanoparticles inhibit Staphylococcus aureus growth. Int. J. Nanomed. 2011, 6, 1553–1558. [Google Scholar]
- Oremland, R.S.; Herbel, M.J.; Blum, J.S.; Langley, S.; Beveridge, T.J.; Ajayan, P.M.; Sutto, T.; Ellis, A.V.; Curran, S. Structural and spectral features of selenium nanospheres produced by se-respiring bacteria. Appl. Environ. Microbiol. 2004, 70, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Matassa, S.; Singh, S.; van Hullebusch, E.D.; Esposito, G.; Lens, P.N.L. Reduction of selenite to elemental selenium nanoparticles by activated sludge. Environ. Sci. Pollut. Res. 2016, 23, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, D. Significance and Roles of Proteus spp. Bacteria in Natural Environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Filali, B.K.; Taoufik, J.; Zeroual, Y.; Dzairi, F.Z.; Talbi, M.; Blaghen, M. Waste water bacterial isolates resistant to heavy metals and antibiotics. Curr. Microbiol. 2000, 41, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.C.; Huang, W.T.; Wu, J.Y.; Houng, J.Y. Microbial decolorization of azo dyes by Proteus mirabilis. J. Ind. Microbiol. Biotechnol. 1999, 23, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Colman, D.R.; Toolson, E.C.; Takacs-Vesbach, C.D. Do diet and taxonomy influence insect gut bacterial communities? Mol. Ecol. 2012, 21, 5124–5137. [Google Scholar] [CrossRef] [PubMed]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef] [PubMed]
- Slobodkina, G.B.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Reduction of chromate, selenite, tellurite, and iron (III) by the moderately thermophilic bacterium Bacillus thermoamylovorans SKC1. Microbiology 2007, 76, 530–534. [Google Scholar] [CrossRef]
- Hunter, W.J.; Kuykendall, L.D. Reduction of selenite to elemental red selenium by Rhizobium sp. strain B1. Curr. Microbiol. 2007, 55, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Rong, Y.; Rui, W.; Dan, W.; Wang, G.; Zheng, S. Reduction of selenite to Se(0) nanoparticles by filamentous bacterium Streptomyces sp. ES2-5 isolated from a selenium mining soil. Microb. Cell Fact. 2016, 15, 157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ren, B.; Li, H.; Lin, Z.; Bañuelos, G.; Li, L.; Zhao, G.; Guo, Y. Biosynthesis of selenium nanoparticles and effects of selenite, selenate, and selenomethionine on cell growth and morphology in Rahnella aquatilis HX2. Appl. Microbiol. Biotechnol. 2018, 102, 6191–6205. [Google Scholar] [CrossRef] [PubMed]
- Khoei, N.S.; Lampis, S.; Zonaro, E.; Yrjälä, K.; Bernardi, P.; Vallini, G. Insights into selenite reduction and biogenesis of elemental selenium nanoparticles by two environmental isolates of Burkholderia fungorum. New Biotechnol. 2017, 34, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Hazard. Mater. 2017, 324, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shu, X.; Zhou, Q.; Fan, T.; Wang, T.; Chen, X.; Li, M.; Ma, Y.; Ni, J.; Hou, J.; et al. Selenite reduction and the biogenesis of selenium nanoparticles by Alcaligenes faecalis Se03 isolated from the gut of Monochamus alternatus (Coleoptera: Cerambycidae). Int. J. Mol. Sci. 2018, 19, 2799. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Błażejak, S. Speciation Analysis of Selenium in Candida utilis Yeast Cells Using HPLC-ICP-MS and UHPLC-ESI-Orbitrap MS Techniques. Appl. Sci. 2018, 8, 2050. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Płaczek, M. Spectrophotometric evaluation of selenium binding by Saccharomyces cerevisiae ATCC MYA-2200 and Candida utilis ATCC 9950 yeast. J. Trace Element Med. Biol. 2016, 35, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Pearce, C.I.; Pattrick, R.A.D.; Nicholas, L.; Charnock, J.M.; Coker, V.S.; Fellowes, J.W.; Oremland, R.S.; Lloyd, J.R. Investigating different mechanisms for biogenic selenite transformations: Geobacter sulfurreducens, Shewanella oneidensis and Veillonella atypica. Environ. Technol. 2009, 30, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Van Fleet-Stalder, V.; Chasteen, T.G.; Pickering, I.J.; George, G.N.; Prince, R.C. Fate of selenate and selenite metabolized by Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2000, 66, 4849–4853. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Bzduchawróbel, A.; Kurcz, A. Effects of Selenium on Morphological Changes in Candida utilis ATCC 9950 Yeast Cells. Biol. Trace Element Res. 2016, 169, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Debieux, C.M.; Dridge, E.J.; Mueller, C.M.; Splatt, P.; Paszkiewicz, K.; Knight, I.; Florance, H.; Love, J.; Titball, R.W.; Lewis, R.J.; et al. A bacterial process for selenium nanosphere assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 13480–13485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobias, J.; Suvorova, E.I.; Bernier-Latmani, R. Role of proteins in controlling selenium nanoparticle size. Nanotechnology 2011, 22, 195605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledgham, F.; Quest, B.; Vallaeys, T.; Mergeay, M.; Covès, J. A probable link between the DedA protein and resistance to selenite. Res. Microbiol. 2005, 156, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Yong, J.; Zhou, H.; Wu, Y. Biosynthesis of Selenium Nanoparticles by a Heavy Metal-Resistant Bacterium Stenotrophomonas sp. EGS12. Nanosci. Nanotechnol. Lett. 2018, 10, 982–987. [Google Scholar] [CrossRef]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [PubMed]
- Ni, T.W.; Staicu, L.C.; Nemeth, R.S.; Schwartz, C.L.; Crawford, D.; Seligman, J.D.; Hunter, W.J.; Pilonsmits, E.A.; Ackerson, C.J. Progress toward clonable inorganic nanoparticles. Nanoscale 2015, 7, 17320–17327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockin, S.L.; Gadd, G.M. Linked redox precipitation of sulfur and selenium under anaerobic conditions by sulfate-reducing bacterial biofilms. Appl. Environ. Microbiol. 2003, 69, 7063–7072. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J. A Rhizobium selenitireducens Protein Showing Selenite Reductase Activity. Curr. Microbiol. 2014, 68, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Wu, S.; Li, N.; Wang, D.; Zheng, S.; Wang, G. Novel bacterial selenite reductase CsrF responsible for Se(IV) and Cr(VI) reduction that produces nanoparticles in Alishewanella sp. WH16-1. J. Hazard. Mater. 2018, 342, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Bébien, M.; Lagniel, G.; Garin, J.; Touati, D.; Verméglio, A.; Labarre, J. Involvement of Superoxide Dismutases in the Response of Escherichia coli to Selenium Oxides. J. Bacteriol. 2002, 184, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Kessi, J. Enzymic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology 2006, 152, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J. Pseudomonas seleniipraecipitans proteins potentially involved in selenite reduction. Curr. Microbiol. 2014, 69, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Sheng, P.; Zhang, H. Isolation and Identification of Cellulolytic Bacteria from the Gut of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae). Int. J. Mol. Sci. 2012, 13, 2563. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, F.; Ahmad, R.; Ashraf, M.Y.; Waraich, E.A.; Khan, S.Z. Effect of selenium foliar spray on physiological and biochemical processes and chemical constituents of wheat under drought stress. Ecotoxicol. Environ. Saf. 2015, 113, 191–200. [Google Scholar] [CrossRef] [PubMed]









| Characteristic | Result | Characteristic | Result |
|---|---|---|---|
| Gram-staining | − | Enzyme activities: | |
| Nitrite reduction | + | α-Glucosidase | + |
| Motility | + | Protease | − |
| Oxidase | − | Utilization of: | |
| Catalase | + | Maltose | − |
| Indole test | + | Lactose | − |
| Nitrate reduction | + | Methyl β-D-glucoside | − |
| Urease | + | cis-Aconitic acid | + |
| Hydrolysis of: | Raffinose | − | |
| Starch | − | L-Arabinose | + |
| Gelatin | + | L-Histidine | + |
| Hydrogen sulfide test | + | Trehalose | + |
| Target Gene | Primer Sequence | Product Size (bp) |
|---|---|---|
| Glutathione synthetase (A0A1Z1SXV1) | Forward 5′-CACGCCAAGTAGCACCT-3′ Reverse 5′-TCAATCTCACGAGCACAA-3′ | 118 |
| Thioredoxin reductase (B4EV88) | Forward 5′-AACCGACCTTTCCGCCTAT-3′ Reverse 5′-CCACCACCGACAACAGCA-3′ | 191 |
| Nitrite reductase (NAD(P)H) (A0A205JSA1) | Forward 5′-GCAAATCGCTCAAGAATA-3′ Reverse 5′-CACCAACTACTGCCTACA-3′ | 186 |
| Glutathione reductase (B4EZ75) | Forward 5′-TAAATGCGTTAGGGAGTG-3′ Reverse 5′-CTGTAGCAGGTTCACGAC-3′ | 246 |
| Fumarate reductase subunit D (B4EWY4) | Forward 5′-CAGGTGGTATGTGGAGTG-3′ Reverse 5′-AAATCGTGCAACGTATGG-3′ | 208 |
| Fumarate reductase subunit C (B4EWY5) | Forward 5′-AACTGGTGGACGAAACTC-3′ Reverse 5′-GCGATAAGGGTCACAATA-3′ | 194 |
| Thioredoxin (B4F1V4) | Forward 5′-CGTGCTCGTTGATTTCT-3′ Reverse 5′-GGTGCTGTCGCAGGGTT-3′ | 138 |
| Sulfite reductase [NADPH] flavoprotein alpha-component (B4F235) | Forward 5′-ATTATCCCGCCACGAAGA-3′ Reverse 5′-AAGCGATGGAGTAAAGACG -3′ | 173 |
| 16S r RNA | Forward 5′-AGAGTTTGATCCTGGCTCAG-3′ Reverse 5′-CTGCTGCCTCCCGTAGGAGT-3′ | 330 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Shu, X.; Hou, J.; Lu, W.; Zhao, W.; Huang, S.; Wu, L. Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification. Int. J. Mol. Sci. 2018, 19, 3809. https://doi.org/10.3390/ijms19123809
Wang Y, Shu X, Hou J, Lu W, Zhao W, Huang S, Wu L. Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification. International Journal of Molecular Sciences. 2018; 19(12):3809. https://doi.org/10.3390/ijms19123809
Chicago/Turabian StyleWang, Yuting, Xian Shu, Jinyan Hou, Weili Lu, Weiwei Zhao, Shengwei Huang, and Lifang Wu. 2018. "Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification" International Journal of Molecular Sciences 19, no. 12: 3809. https://doi.org/10.3390/ijms19123809
APA StyleWang, Y., Shu, X., Hou, J., Lu, W., Zhao, W., Huang, S., & Wu, L. (2018). Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification. International Journal of Molecular Sciences, 19(12), 3809. https://doi.org/10.3390/ijms19123809