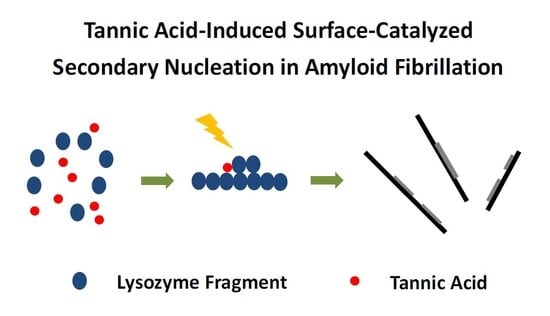
Tannic Acid-Induced Surface-Catalyzed Secondary Nucleation during the Amyloid Fibrillation of Hen Egg-White Lysozyme
Abstract
:1. Introduction
2. Results and Discussion
2.1. ThT Fluorescence Assay
2.2. AFM Investigation
2.3. Fluorescence Quenching Investigation at 323, 328, and 333 K.
2.3.1. Fluorescence Quenching Spectra of the Lysozyme–Tannic Acid System
2.3.2. Association Constants and Number of Binding Sites
2.3.3. Determination of Binding Forces
2.2.4. Binding Distances
2.2.5. Fluorescence Quenching Investigation at 298, 303, and 308 K.
3. Materials and Methods
3.1. Materials
3.2. Amyloid Fibrillation
3.3. AFM
3.4. ThT Fluorescence Assay
3.5. UV-Vis Measurement
3.6. Fluorescence Quenching Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nelson, R.; Eisenberg, D. Structural models of amyloid-like fibrils. Adv. Protein Chem. 2006, 73, 235–282. [Google Scholar] [PubMed]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.W.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Jucker, M. The amyloid state of proteins in human diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.D.; Cohen, A.S. Review: History of the amyloid fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Klunk, W.E.; Jacob, R.F.; Mason, R.P. Quantifying amyloid by Congo red spectral shift assay. Methods Enzymol. 1999, 309, 285–305. [Google Scholar]
- LeVine, H., 3rd. Quantification of beta-sheet amyloid fibril structures with thioflavin t. Methods Enzymol. 1999, 309, 274–284. [Google Scholar] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Hauser, C.A.E.; Maurer-Stroh, S.; Martins, I.C. Amyloid-based nanosensors and nanodevices. Chem. Soc. Rev. 2014, 43, 5326–5345. [Google Scholar] [CrossRef] [PubMed]
- Shimanovich, U.; Efimov, I.; Mason, T.O.; Flagmeier, P.; Buell, A.K.; Gedanken, A.; Linse, S.; Akerfeldt, K.S.; Dobson, C.M.; Weitz, D.A.; et al. Protein microgels from amyloid fibril networks. ACS Nano 2015, 9, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Li, Y.F.; Zhong, C. Amyloid-directed assembly of nanostructures and functional devices for bionanoelectronics. J. Mater. Chem. B 2015, 3, 4953–4958. [Google Scholar] [CrossRef]
- Li, C.X.; Adamcik, J.; Mezzenga, R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat. Nanotechnol. 2012, 7, 421–427. [Google Scholar] [CrossRef]
- Reynolds, N.P.; Charnley, M.; Mezzenga, R.; Hartley, P.G. Engineered lysozyme amyloid fibril networks support cellular growth and spreading. Biomacromolecules 2014, 15, 599–608. [Google Scholar] [CrossRef]
- Cohen, S.I.A.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012, 421, 160–171. [Google Scholar] [CrossRef]
- Jeong, J.S.; Ansaloni, A.; Mezzenga, R.; Lashuel, H.A.; Dietler, G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J. Mol. Biol. 2013, 425, 1765–1781. [Google Scholar] [CrossRef]
- Mansson, C.; Arosio, P.; Hussein, R.; Kampinga, H.H.; Hashem, R.M.; Boelens, W.C.; Dobson, C.M.; Knowles, T.P.J.; Linse, S.; Emanuelsson, C. Interaction of the molecular chaperone DNAJB6 with growing arnyloid-beta 42 (Abeta 42) aggregates leads to sub-stoichiometric inhibition of amyloid formation. J. Biol. Chem. 2014, 289, 31066–31076. [Google Scholar] [CrossRef]
- Abelein, A.; Graslund, A.; Danielsson, J. Zinc as chaperone-mimicking agent for retardation of amyloid beta peptide fibril formation. Proc. Natl. Acad. Sci. USA 2015, 112, 5407–5412. [Google Scholar] [CrossRef]
- Abelein, A.; Jarvet, J.; Barth, A.; Graslund, A.; Danielsson, J. Ionic strength modulation of the free energy landscape of Abeta(40) peptide fibril formation. J. Am. Chem. Soc. 2016, 138, 6893–6902. [Google Scholar] [CrossRef]
- Krebs, M.R.H.; Wilkins, D.K.; Chung, E.W.; Pitkeathly, M.C.; Chamberlain, A.K.; Zurdo, J.; Robinson, C.V.; Dobson, C.M. Formation and seeding of amyloid fibrils from wild-type hen lysozyme and a peptide fragment from the beta-domain. J. Mol. Biol. 2000, 300, 541–549. [Google Scholar] [CrossRef]
- Frare, E.; de Laureto, P.P.; Zurdo, J.; Dobson, C.M.; Fontana, A. A highly amyloidogenic region of hen lysozyme. J. Mol. Biol. 2004, 340, 1153–1165. [Google Scholar] [CrossRef]
- Arnaudov, L.N.; de Vries, R. Thermally induced fibrillar aggregation of hen egg white lysozyme. Biophys. J. 2005, 88, 515–526. [Google Scholar] [CrossRef]
- Mishra, R.; Sorgjerd, K.; Nystrom, S.; Nordigarden, A.; Yu, Y.-C.; Hammarstrom, P. Lysozyme amyloidogenesis is accelerated by specific nicking and fragmentation but decelerated by intact protein binding and conversion. J. Mol. Biol. 2007, 366, 1029–1044. [Google Scholar] [CrossRef]
- Hill, S.E.; Robinson, J.; Matthews, G.; Muschol, M. Amyloid protofibrils of lysozyme nucleate and grow via oligomer fusion. Biophys. J. 2009, 96, 3781–3790. [Google Scholar] [CrossRef]
- Kumar, S.; Ravi, V.K.; Swaminathan, R. How do surfactants and dtt affect the size, dynamics, activity and growth of soluble lysozyme aggregates? Biochem. J. 2008, 415, 275–288. [Google Scholar] [CrossRef]
- Goda, S.; Takano, K.; Yamagata, Y.; Nagata, R.; Akutsu, H.; Maki, S.; Namba, K.; Yutani, K. Amyloid protofilament formation of hen egg lysozyme in highly concentrated ethanol solution. Protein Sci. 2000, 9, 369–375. [Google Scholar] [CrossRef]
- Khan, J.M.; Qadeer, A.; Chaturvedi, S.K.; Ahmad, E.; Rehman, S.A.; Gourinath, S.; Khan, R.H. SDS can be utilized as an amyloid inducer: A case study on diverse proteins. PLoS ONE 2012, 7, e29694. [Google Scholar] [CrossRef]
- Vernaglia, B.A.; Huang, J.; Clark, E.D. Guanidine hydrochloride can induce amyloid fibril formation from hen egg-white lysozyme. Biomacromolecules 2004, 5, 1362–1370. [Google Scholar] [CrossRef]
- Sasahara, K.; Yagi, H.; Naiki, H.; Goto, Y. Heat-induced conversion of beta(2)-microglobulin and hen egg-white lysozyme into amyloid fibrils. J. Mol. Biol. 2007, 372, 981–991. [Google Scholar] [CrossRef]
- Takase, K.; Higashi, T.; Omura, T. Aggregate formation and the structure of the aggregates of disulfide-reduced proteins. J. Protein Chem. 2002, 21, 427–433. [Google Scholar] [CrossRef]
- Zou, Y.; Li, Y.; Hao, W.; Hu, X.; Ma, G. Parallel beta-sheet fibril and antiparallel beta-sheet oligomer: New insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from ftir spectroscopy. J. Phys. Chem. B 2013, 117, 4003–4013. [Google Scholar] [CrossRef]
- Zou, Y.; Hao, W.; Li, H.; Gao, Y.; Sun, Y.; Ma, G. New insight into amyloid fibril formation of hen egg white lysozyme using a two-step temperature-dependent ftir approach. J. Phys. Chem. B 2014, 118, 9834–9843. [Google Scholar] [CrossRef]
- Mohammadi, F.; Moeeni, M.; Mahmudian, A.; Hassani, L. Inhibition of amyloid fibrillation of lysozyme by bisdemethoxycurcumin and diacetylbisdemethoxycurcumin. Biophys. Chem. 2018, 235, 56–65. [Google Scholar] [CrossRef]
- Seraj, Z.; Seyedarabi, A.; Saboury, A.A.; Habibi-Rezaei, M.; Ahmadian, S.; Ghasemi, A. Unraveling the novel effects of aroma from small molecules in preventing hen egg white lysozyme amyloid fibril formation. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Patel, P.; Parmar, K.; Patel, D.; Kumar, S.; Trivedi, M.; Das, M. Inhibition of amyloid fibril formation of lysozyme by ascorbic acid and a probable mechanism of action. Int. J. Biol. Macromol. 2018, 114, 666–678. [Google Scholar] [CrossRef]
- How, S.-C.; Cheng, Y.-H.; Lo, C.-H.; Lai, J.-T.; Lin, T.-H.; Bednarikova, Z.; Antosova, A.; Gazova, Z.; Wu, J.W.; Wang, S.S.S. Exploring the effects of methylene blue on amyloid fibrillogenesis of lysozyme. Int. J. Biol. Macromol. 2018, 119, 1059–1067. [Google Scholar] [CrossRef]
- Porat, Y.; Abramowitz, A.; Gazit, E. Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug Des. 2006, 67, 27–37. [Google Scholar] [CrossRef]
- Ono, K.; Yamada, M. Antioxidant compounds have potent anti-fibrillogenic and fibril-destabilizing effects for alpha-synuclein fibrils in vitro. J. Neurochem. 2006, 97, 105–115. [Google Scholar] [CrossRef] [Green Version]
- Boye-Harnasch, M.; Cullin, C. A novel in vitro filter trap assay identifies tannic acid as an amyloid aggregation inducer for HET-s. J. Biotechnol. 2006, 125, 222–230. [Google Scholar] [CrossRef]
- Khan, M.V.; Ishtikhar, M.; Siddiqui, M.K.; Zaman, M.; Chandel, T.I.; Majid, N.; Ajmal, M.R.; Abdelhameed, A.S.; Shahein, Y.E.; Khan, R.H. Biophysical insight reveals tannic acid as amyloid inducer and conformation transformer from amorphous to amyloid aggregates in concanavalin A (ConA). J. Biomol. Struct. Dyn. 2018, 36, 1261–1273. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochimica Et Biophysica Acta Proteins Proteom. 2010, 1804, 1405–1412. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Interaction of thioflavin T with amyloid fibrils: Stoichiometry and affinity of dye binding, absorption spectra of bound dye. J. Phys. Chem. B 2011, 115, 11519–11524. [Google Scholar] [CrossRef]
- Ghisaidoobe, A.B.T.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on forster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006. [Google Scholar]
- Cao, S.N.; Liu, B.S.; Li, Z.Y.; Chong, B.H. A fluorescence spectroscopic study of the interaction between glipizide and bovine serum albumin and its analytical application. J. Lumin. 2014, 145, 94–99. [Google Scholar] [CrossRef]
- Shanmugaraj, K.; Anandakumar, S.; Ilanchelian, M. Probing the binding interaction of thionine with lysozyme: A spectroscopic and molecular docking investigation. Dyes Pigm. 2015, 112, 210–219. [Google Scholar] [CrossRef]
- Xiong, X.; He, J.; Yang, H.; Tang, P.; Tang, B.; Sun, Q.; Li, H. Investigation on the interaction of antibacterial drug moxifloxacin hydrochloride with human serum albumin using multi-spectroscopic approaches, molecular docking and dynamical simulation. RSC Adv. 2017, 7, 48942–48951. [Google Scholar] [CrossRef] [Green Version]
- Fonin, A.V.; Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Formoso, C.; Forster, L.S. Tryptophan fluorescence lifetimes in lysozyme. J. Biol. Chem. 1975, 250, 3738–3745. [Google Scholar]
- Li, D.; Cao, X.; Ji, B. Spectrophotometric studies on the interaction between myricetin and lysozyme in the absence or presence of Cu2+ or Fe3+. J. Lumin. 2010, 130, 1893–1900. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Mohammadi, F.; Mahmudian, A.; Moeeni, M.; Hassani, L. Inhibition of amyloid fibrillation of hen egg-white lysozyme by the natural and synthetic curcuminoids. RSC Adv. 2016, 6, 23148–23160. [Google Scholar] [CrossRef]
- Powers, E.T.; Powers, D.L. Mechanisms of protein fibril formation: Nucleated polymerization with competing off-pathway aggregation. Biophys. J. 2008, 94, 379–391. [Google Scholar] [CrossRef]








| T/K | Ksv/(L·mol−1) | Kq/(L·mol−1·s−1) | r * |
|---|---|---|---|
| 323 | (1.30 ± 0.10) × 104 | (1.44 ± 0.11) × 1012 | 0.9948 ± 0.0024 |
| 328 | (1.01 ± 0.09) × 104 | (1.12 ± 0.10) × 1012 | 0.9926 ± 0.0030 |
| 333 | ((0.97 ± 0.11) × 104 | (1.08 ± 0.12) × 1012 | 0.9915 ± 0.0044 |
| T/K | KA/(L·mol−1) | n | r * |
|---|---|---|---|
| 323 | (6.83 ± 4.09) × 103 | 0.91 ± 0.09 | 0.9901 ± 0.0051 |
| 328 | (2.16 ± 1.59) × 104 | 1.07 ± 0.09 | 0.9943 ± 0.0016 |
| 333 | (4.24 ± 4.02) × 104 | 1.13 ± 0.09 | 0.9968 ± 0.0020 |
| T/K | ΔH/(kJ·mol−1) | r * | ΔG/(kJ·mol−1) | ΔS/[J·(mol·K−1)−1] |
|---|---|---|---|---|
| 323 | 139.38 | 0.9901 | −21.88 | 541.13 |
| 328 | −25.14 | 542.99 | ||
| 333 | −27.28 | 541.11 |
| T/K | E/% | J/(cm3·L·mol−1) | R0/nm | r/nm |
|---|---|---|---|---|
| 323 | 32.45 ± 0.35 | (5.95 ± 0.04) × 10−15 | 2.34 ± 2.48 × 10−3 | 2.64 ± 0.01 |
| 328 | 23.51 ± 3.88 | (6.00 ± 0.04) × 10−15 | 2.34 ± 2.78 × 10−3 | 2.86 ± 0.11 |
| 333 | 22.95 ± 1.55 | (6.15 ± 0.26) × 10−15 | 2.34 ± 1.63 × 10−2 | 2.88 ± 0.02 |
| T/K | ΔH/(kJ·mol−1) | r * | ΔG/(kJ·mol−1) | ΔS/[J·(mol·K−1)−1] |
|---|---|---|---|---|
| 298 | −97.28 | 0.9999 | −28.92 | −229.42 |
| 303 | −27.80 | −229.30 | ||
| 308 | −26.62 | −229.42 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, J.; Yu, Y.; Wang, Y.; Li, H.; Yang, L.; Du, B.; Ma, G. Tannic Acid-Induced Surface-Catalyzed Secondary Nucleation during the Amyloid Fibrillation of Hen Egg-White Lysozyme. Int. J. Mol. Sci. 2018, 19, 4009. https://doi.org/10.3390/ijms19124009
Tian J, Yu Y, Wang Y, Li H, Yang L, Du B, Ma G. Tannic Acid-Induced Surface-Catalyzed Secondary Nucleation during the Amyloid Fibrillation of Hen Egg-White Lysozyme. International Journal of Molecular Sciences. 2018; 19(12):4009. https://doi.org/10.3390/ijms19124009
Chicago/Turabian StyleTian, Jing, Yang Yu, Yao Wang, Haoyi Li, Lujuan Yang, Baoan Du, and Gang Ma. 2018. "Tannic Acid-Induced Surface-Catalyzed Secondary Nucleation during the Amyloid Fibrillation of Hen Egg-White Lysozyme" International Journal of Molecular Sciences 19, no. 12: 4009. https://doi.org/10.3390/ijms19124009
APA StyleTian, J., Yu, Y., Wang, Y., Li, H., Yang, L., Du, B., & Ma, G. (2018). Tannic Acid-Induced Surface-Catalyzed Secondary Nucleation during the Amyloid Fibrillation of Hen Egg-White Lysozyme. International Journal of Molecular Sciences, 19(12), 4009. https://doi.org/10.3390/ijms19124009