Physicochemical Properties of the Mammalian Molecular Chaperone HSP60
Abstract
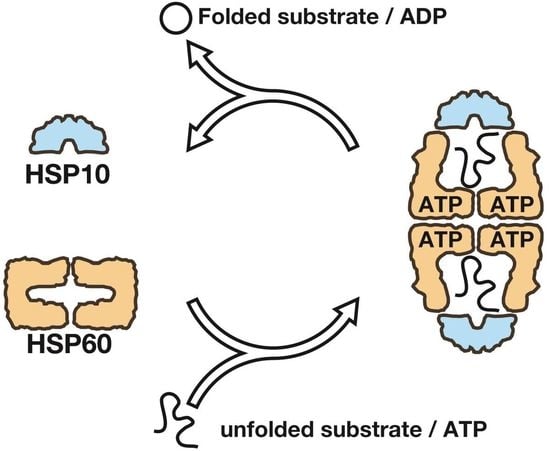
:1. Introduction
2. Results
2.1. The Double Ring Formation of HSP60 Depends on the Nucleotide Conditions and HSP10
2.2. The Association of the HSP60 Single Rings Was Induced by the Presence of ATP and Increased by the Addition of HSP10
2.3. The Association between HSP60 and HSP10 Was Induced by Only ATP
2.4. ATP Binding State of HSP60 Stably Interacts with HSP10 and Forms a Football-Type Complex
2.5. HSP60/HSP10 Complex Dissociates after ATP Was Hydrolyzed
3. Discussion
4. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Pechmann, S.; Willmund, F.; Frydman, J. The ribosome as a hub for protein quality control. Mol. Cell 2013, 49, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Saibil, H.R.; Fenton, W.A.; Clare, D.K.; Horwich, A.L. Structure and allostery of the chaperonin GroEL. J. Mol. Biol. 2013, 425, 1476–1487. [Google Scholar] [CrossRef] [PubMed]
- Braig, K.; Otwinowski, Z.; Hegde, R.; Boisvert, D.C.; Joachimiak, A.; Horwich, A.L.; Sigler, P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 1994, 371, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Horwich, A.L.; Sigler, P.B. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 1997, 388, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.Y.; Hartl, F.U.; Martin, J.; Pollock, R.A.; Kalousek, F.; Neupert, W.; Hallberg, E.M.; Hallberg, R.L.; Horwich, A.L. Mitochondrial heat-shock protein HSP60 is essential for assembly of proteins imported into yeast mitochondria. Nature 1989, 337, 620–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostermann, J.; Horwich, A.L.; Neupert, W.; Hartl, F.U. Protein folding in mitochondria requires complex formation with HSP60 and ATP hydrolysis. Nature 1989, 341, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manning-Krieg, U.C.; Scherer, P.E.; Schatz, G. Sequential action of mitochondrial chaperones in protein import into the matrix. EMBO J. 1991, 10, 3273–3280. [Google Scholar] [PubMed]
- Koll, H.; Guiard, B.; Rassow, J.; Osterman, J.; Horwich, A.L.; Neupert, W.; Hartl, F.U. Antifolding Activity of HSP60 couples Protein Import into the Mitochondrial Matrix with Export to the Intermembrane Space. Cell 1992, 68, 1163–1175. [Google Scholar] [CrossRef]
- Rospert, S.; Looser, R.; Dubaquie, Y.; Matouschek, A.; Glick, B.S.; Schatz, G. HSP60 independent protein folding in the matrix of yeast mitochondria. EMBO J. 1996, 15, 764–774. [Google Scholar] [PubMed]
- Hallberg, E.M.; Shu, Y.; Hallberg, R.L. Loss of mitochondrial HSP60 function: Nonequivelent effects on matrix-targeted and intermembrane-targeted proteins. Mol. Cell. Biol. 1993, 13, 3050–3057. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Komatsuda, A.; Ohtani, H.; Wakui, H.; Imai, H.; Sawada, K.; Otaka, M.; Ogura, M.; Suzuki, A.; Hamada, F. Mammalian HSP60 is quickly sorted into the mitochondria under conditions of dehydration. Eur. J. Biochem. 2002, 269, 5931–5938. [Google Scholar] [PubMed]
- Itoh, H.; Kobayashi, R.; Wakui, H.; Komatsuda, A.; Ohtani, H.; Miura, A.B.; Otaka, M.; Masamune, O.; Andoh, H.; Koyama, K.; et al. Mammalian 60-kDa stress protein (chaperonin homolog). Identification, biochemical properties, and localization. J. Biol. Chem. 1995, 270, 13429–13435. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Komatsuda, A.; Wakui, H.; Miura, A.B.; Tashima, Y. Mammalian HSP60 is a major target for an immunosuppressant mizoribine. J. Biol. Chem. 1999, 274, 35147–35151. [Google Scholar] [CrossRef] [PubMed]
- Viitanen, P.V.; Lorimer, G.H.; Seetharam, R.; Gupta, R.S.; Oppenheim, J.; Thomas, J.O.; Cowan, N.J. Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J. Biol. Chem. 1992, 267, 695–698. [Google Scholar] [PubMed]
- Nielsen, K.L.; Cowan, N.J. A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol. Cell 1998, 2, 93–99. [Google Scholar] [CrossRef]
- Viitanen, P.V.; Lorimer, G.; Bergmeier, W.; Weiss, C.; Kessel, M.; Goloubinoff, P. Purification of mammalian mitochondrial chaperonin 60 through in vitro reconstitution of active oligomers. Methods Enzymol. 1998, 290, 203–217. [Google Scholar] [PubMed]
- Levy-Rimler, G.; Viitanen, P.; Weiss, C.; Sharkia, R.; Greenberg, A.; Niv, A.; Lustig, A.; Delarea, Y.; Azem, A. The effect of nucleotides and mitochondrial chaperonin 10 on the structure and chaperone activity of mitochondrial chaperonin 60. Eur. J. Biochem. 2001, 268, 3465–3472. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Ishida, R.; Yamamoto, H.; Tanabe-Ishida, M.; Haga, A.; Takahashi, H.; Takahashi, K.; Goto, D.; Grave, E.; Itoh, H. Functional structure and physiological functions of mammalian wild-type HSP60. Arch. Biochem. Biophys. 2015, 586, 10–19. [Google Scholar]
- Nisemblat, S.; Yaniv, O.; Parnas, A.; Frolow, F.; Azem, A. Crystal structure of the human mitochondrial chaperonin symmetrical football complex. Proc. Natl. Acad. Sci. USA 2015, 112, 6044–6049. [Google Scholar] [CrossRef] [PubMed]
- Parnas, A.; Nisemblat, S.; Weiss, C.; Levy-Rimler, G.; Pri-Or, A.; Zor, T.; Lund, P.A.; Bross, P.; Azem, A. Identification of elements that dictate the specificity of mitochondrial Hsp60 for its co-chaperonin. PLoS ONE 2012, 7, e50318. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Song, J.L.; Chuang, D.T.; Chiu, W.; Ludtke, S.J. An expanded conformation of single-ring GroEL-GroES complex encapsulates an 86 kDa substrate. Structure 2006, 14, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.; Hayer-Hartl, M.K.; Goldie, K.N.; Pfeifer, G.; Hegerl, R.; Müller, S.; da Silva, A.C.; Baumeister, W.; Hartl, F.U. Functional significance of symmetrical versus asymmetrical GroEL-GroES chaperonin complexes. Science 1995, 269, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Inobe, T.; Takahashi, K.; Maki, K.; Enoki, S.; Kamagata, K.; Kadooka, A.; Arai, M.; Kuwajima, K. Asymmetry of the GroEL-GroES complex under physiological conditions as revealed by small-angle X-ray scattering. Biophys. J. 2008, 94, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Bacia, K.; Schwille, P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2007, 2, 2842–2856. [Google Scholar] [CrossRef] [PubMed]
- Muto, H.; Kinjo, M.; Yamamoto, K.T. Fluorescence cross-correlation spectroscopy of plant proteins. Methods Mol. Biol. 2009, 479, 203–215. [Google Scholar] [PubMed]
- Rye, H.S.; Burston, S.G.; Fenton, W.A.; Beechem, J.M.; Xu, Z.; Sigler, P.B.; Horwich, A.L. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 1997, 388, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Koike-Takeshita, A.; Yoshida, M.; Taguchi, H. Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J. Biol. Chem. 2008, 283, 23774–23781. [Google Scholar] [CrossRef] [PubMed]
- Motojima, F.; Yoshida, M. Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL: Hexokinase treatment revealed the exclusive role of ATP. J. Biol. Chem. 2003, 278, 26648–26654. [Google Scholar] [CrossRef] [PubMed]
- Hayer-Hartl, M.K.; Martin, J.; Hartl, F.U. Asymmetrical interaction of GroEL and GroES in the ATPase cycle of assisted protein folding. Science 1995, 269, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Weissman, J.S.; Rye, H.S.; Fenton, W.A.; Beechem, J.M.; Horwich, A.L. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell 1996, 84, 481–490. [Google Scholar] [CrossRef]
- Rye, H.S.; Roseman, A.M.; Chen, S.; Furtak, K.; Fenton, W.A.; Saibil, H.R.; Horwich, A.L. GroEL-GroES cycling: ATP and nonnative polypeptide direct alternation of folding-active rings. Cell 1999, 97, 325–338. [Google Scholar] [CrossRef]
- Ueno, T.; Taguchi, H.; Tadakuma, H.; Yoshida, M.; Funatsu, T. GroEL mediates protein folding with a two successive timer mechanism. Mol. Cell 2004, 14, 423–434. [Google Scholar] [CrossRef]
- Sameshima, T.; Ueno, T.; Iizuka, R.; Ishii, N.; Terada, N.; Okabe, K.; Funatsu, T. Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J. Biol. Chem. 2008, 283, 23765–23773. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Rutkat, K.; Rachel, R.; Pfeifer, G.; Jaenicke, R.; Viitanen, P.; Lorimer, G.; Buchner, J. Symmetric complexes of GroE chaperonins as part of the functional cycle. Science 1994, 265, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Azem, A.; Diamant, S.; Kessel, M.; Weiss, C.; Goloubinoff, P. The protein-folding activity of chaperonins correlates with the symmetric GroEL14 (GroES7)2 heterooligomer. Proc. Natl. Acad. Sci. USA 1995, 92, 12021–12025. [Google Scholar] [CrossRef] [PubMed]
- Azem, A.; Kessel, M.; Goloubinoff, P. Characterization of a functional GroEL14 (GroES7)2 chaperonin hetero-oligomer. Science 1994, 265, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zvi, A.P.; Chatellier, J.; Fersht, A.R.; Goloubinoff, P. Minimal and optimal mechanisms for GroE-mediated protein folding. Proc. Natl. Acad. Sci. USA 1998, 95, 15275–15280. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, H.; Tsukuda, K.; Motojima, F.; Koike-Takeshita, A.; Yoshida, M. BeF(x) stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J. Biol. Chem. 2004, 279, 45737–45743. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Ye, X.; Lorimer, G.H. Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci. USA 2013, 110, E4298–E4305. [Google Scholar] [CrossRef] [PubMed]
- Sameshima, T.; Iizuka, R.; Ueno, T.; Funatsu, T. Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem. J. 2010, 427, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Lorimer, G.H. Substrate protein switches GroE chaperonins from asymmetric to symmetric cycling by catalyzing nucleotide exchange. Proc. Natl. Acad. Sci. USA 2013, 110, E4289–E4297. [Google Scholar] [CrossRef] [PubMed]
- Koike-Takeshita, A.; Mitsuoka, K.; Taguchi, H. Asp-52 in combination with Asp-398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J. Biol. Chem. 2014, 289, 30005–30011. [Google Scholar] [CrossRef] [PubMed]
- Koike-Takeshita, A.; Arakawa, T.; Taguchi, H.; Shimamura, T. Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 Å reveals rearrangement between two GroEL rings. J. Mol. Biol. 2014, 426, 3634–3641. [Google Scholar]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [PubMed]
- Noguchi, S.; Toyoshima, T.; Yamamoto, S.; Miyazaki, T.; Otaka, M.; Watanabe, S.; Imai, K.; Senoo, H.; Kobayashi, R.; Jikei, M.; et al. Cytosolic chaperonin CCT possesses GTPase activity. Am. J. Mol. Biol. 2011, 1, 123–130. [Google Scholar] [CrossRef]







| Nucleotide | RCA | Binding Ratio (%) | Kd (M) |
|---|---|---|---|
| Free | 0.22 | 35.1 | 7.5 × 10−7 |
| ATP | 0.55 | 86.8 | 8.7 × 10−8 |
| ADP | 0.25 | 40.2 | 5.1 × 10−7 |
| AMP-PNP | 0.20 | 32.5 | 7.9 × 10−7 |
| ATPγS | 0.23 | 36.6 | 7.2 × 10−7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishida, R.; Okamoto, T.; Motojima, F.; Kubota, H.; Takahashi, H.; Tanabe, M.; Oka, T.; Kitamura, A.; Kinjo, M.; Yoshida, M.; et al. Physicochemical Properties of the Mammalian Molecular Chaperone HSP60. Int. J. Mol. Sci. 2018, 19, 489. https://doi.org/10.3390/ijms19020489
Ishida R, Okamoto T, Motojima F, Kubota H, Takahashi H, Tanabe M, Oka T, Kitamura A, Kinjo M, Yoshida M, et al. Physicochemical Properties of the Mammalian Molecular Chaperone HSP60. International Journal of Molecular Sciences. 2018; 19(2):489. https://doi.org/10.3390/ijms19020489
Chicago/Turabian StyleIshida, Ryuichi, Tomoya Okamoto, Fumihiro Motojima, Hiroshi Kubota, Hiroki Takahashi, Masako Tanabe, Toshihiko Oka, Akira Kitamura, Masataka Kinjo, Masasuke Yoshida, and et al. 2018. "Physicochemical Properties of the Mammalian Molecular Chaperone HSP60" International Journal of Molecular Sciences 19, no. 2: 489. https://doi.org/10.3390/ijms19020489
APA StyleIshida, R., Okamoto, T., Motojima, F., Kubota, H., Takahashi, H., Tanabe, M., Oka, T., Kitamura, A., Kinjo, M., Yoshida, M., Otaka, M., Grave, E., & Itoh, H. (2018). Physicochemical Properties of the Mammalian Molecular Chaperone HSP60. International Journal of Molecular Sciences, 19(2), 489. https://doi.org/10.3390/ijms19020489