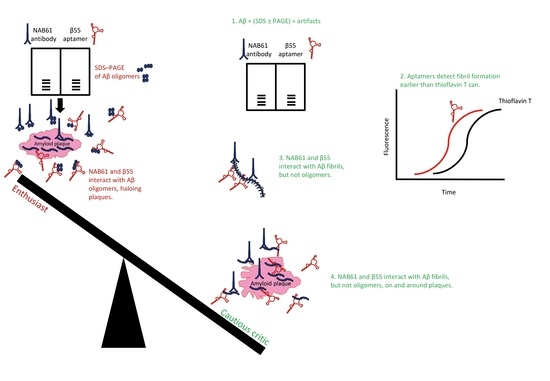
Aptamers Selected for Recognizing Amyloid β-Protein—A Case for Cautious Optimism
Abstract
:1. Introduction
2. Amyloid β-Protein and Alzheimer Disease
3. Aptamers and Systematic Evolution of Ligands by Exponential Enrichment
4. Aptamers and Aβ
5. SDS–PAGE, Aptamers, Antibodies, and “Halos of Oligomers”
6. Conclusions
- The handful of reports published since 2002 on aptamers developed for targeting Aβ have led to important and instructive findings. RNA and DNA aptamers and random nucleotide libraries used for selecting aptamers are found to react inherently and nonspecifically with fibrillar Aβ preparations and exemplary amyloid assemblies [21,153,160]. Most likely, the aptamer-targeted common aptatope in these cases is the backbone of the proteins in a cross-β structure because this protein structure reportedly facilitates retention of RNAs and RNA-binding proteins into special ribonucleoprotein complexes, including stress granules and RNA-processing organelles [189]. The inherent and persistent tendency of RNA aptamers to bind amyloid fibrils (or vice versa) may explain entrapment of RNA in the senile plaques and neurofibrillary tangles [73,74,75], the two pathological hallmarks of AD brains. Moreover, amyloid fibrils and oligonucleotides act as polyelectrolytes and interact by electrostatic forces [190]. These β-sheet-mediated, polyelectrolytic, protein–oligonucleotide interactions were thought to be vital for support, stability, compartmentalization, protection, and resistance to degradation in the harsh environments of the antediluvian, prebiotic world [191], indicating an ancient phenomenon. Interaction of RNA aptamers with amyloid fibrils have implications for the previous and future studies of aptamers selected for amyloidogenic proteins and conclusions drawn from such studies.
- Attributing oligomer specificity to an aptamer based on results obtained by SDS–PAGE fractionation of Aβ preparations disregards the collected evidence on the unsuitability of SDS–PAGE for analyzing and size estimation of amyloidogenic protein assemblies.
- Attributing oligomer specificity to an aptamer (or an antibody) that evidently binds fibrillar structures of amyloidogenic proteins (see [127,163]) is erroneous and misleading; thus, binding specificities of such aptamers in tissue sections do not represent their true specificities and enhances the illusion about presence of Aβ oligomers in tissue sections.
- Finally, I hope this review could encourage the aptamer–amyloid–Alzheimer researchers, the relevant funding bodies, these fields’ peer-reviewers, and the fields’ young scholars to scrutinize and study the relevant literature deeply before enthusing [148,200,201,202] about aptamers in the context of Aβ research. Let us not generate an aptamer field akin to the muddled assortment of antibodies promoted in AD research [21,22].
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | amyloid β-protein |
| AD | Alzheimer disease |
| APP | amyloid β-protein precursor |
| ELISA | enzyme-linked immunosorbent assay |
| ESI–IM–MS | ion mobility coupled with electrospray-ionization mass spectrometry |
| HFIP | 1,1,1,3,3,3-hexafluoro-2-propanol |
| IAPP | islet amyloid polypeptide |
| IDPs | intrinsically disordered proteins |
| IM–MS | ion-mobility spectrometry–mass spectrometery |
| PCR | polymerase chain reaction |
| PICUP | photo-induced crosslinking of unmodified proteins |
| PLGA | poly(lactic-co-glycolic acid) |
| PrP | prion proteins |
| SDS–PAGE | sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SELEX | systematic evolution of ligands by exponential enrichment |
References
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Famulok, M.; Mayer, G. Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol. 1999, 243, 123–136. [Google Scholar] [PubMed]
- Tan, W.; Wang, H.; Chen, Y.; Zhang, X.; Zhu, H.; Yang, C.; Yang, R.; Liu, C. Molecular aptamers for drug delivery. Trends Biotechnol. 2011, 29, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Castanares, M.; Mukherjee, A.; Lupold, S.E. Nucleic acid aptamers: Clinical applications and promising new horizons. Curr. Med. Chem. 2011, 18, 4206–4214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent advances in SELEX technology and aptamer applications in biomedicine. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.A.; Keating, G.M. Pegaptanib: In exudative age-related macular degeneration. Drugs 2005, 65, 1571–1577; discussion 1578–1579. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, A.R.; Brandl, C.J.; Struhl, K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of yeast GCN4 protein. Mol. Cell. Biol. 1989, 9, 2944–2949. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Vater, A.; Klussmann, S. Turning mirror-image oligonucleotides into drugs: The evolution of Spiegelmer® therapeutics. Drug Discov. Today 2015, 20, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: Another illustration of the D2 concept. Expert Rev. Proteom. 2010, 7, 543–564. [Google Scholar] [CrossRef] [PubMed]
- Ambadipudi, S.; Zweckstetter, M. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin. Drug Discov. 2016, 11, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Bahadur, R.P. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell. Mol. Life Sci. 2016, 73, 4075–4084. [Google Scholar] [CrossRef] [PubMed]
- Korsak, M.; Kozyreva, T. Beta Amyloid hallmarks: From intrinsically disordered proteins to Alzheimer’s disease. Adv. Exp. Med. Biol. 2015, 870, 401–421. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, M. Fibril formation from the amyloid-β peptide is governed by a dynamic equilibrium involving association and dissociation of the monomer. Biophys. Rev. 2017, 9, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Sarsoza, F.; Saing, T.; Cotman, C.W.; Necula, M.; Margol, L.; Wu, J.; Breydo, L.; Thompson, J.L.; et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2007, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Bitan, G. Overview of fibrillar and oligomeric assemblies of amyloidogenic proteins. In Non-Fibrillar Amyloidogenic Protein Assemblies—Common Cytotoxins Underlying Degenerative Diseases; Rahimi, F., Bitan, G., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–36. ISBN 978-9-40-072773-1. [Google Scholar]
- Hunter, S.; Brayne, C. Do anti-amyloid beta protein antibody cross reactivities confound Alzheimer disease research? J. Negat. Results Biomed. 2017, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Wolfe, M.S. A portrait of Alzheimer secretases—New features and familiar faces. Science 2001, 293, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.N.; Robinson, P.J.; Chleboun, J.O.; Beyreuther, K.; Masters, C.L. The molecular pathology of amyloid deposition in Alzheimer’s disease. Mol. Neurobiol. 1991, 5, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Hung, A.Y.; Schlossmacher, M.G.; Oltersdorf, T.; Teplow, D.B.; Selkoe, D.J. Normal cellular processing of the β-amyloid precursor protein results in the secretion of the amyloid β peptide and related molecules. Ann. N. Y. Acad. Sci. 1993, 695, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Parihar, M.S.; Brewer, G.J. Amyloid-β as a modulator of synaptic plasticity. J. Alzheimers Dis. 2010, 22, 741–763. [Google Scholar] [CrossRef] [PubMed]
- Puzzo, D.; Arancio, O. Amyloid-β peptide: Dr. Jekyll or Mr. Hyde? J. Alzheimers Dis. 2013, 33, S111–S120. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Mora, P.; Luna, R.; Colín-Barenque, L. Amyloid beta: Multiple mechanisms of toxicity and only some protective effects? Oxid. Med. Cell. Longev. 2014, 2014, 795375. [Google Scholar] [CrossRef] [PubMed]
- Dawkins, E.; Small, D.H. Insights into the physiological function of the β-amyloid precursor protein: Beyond Alzheimer’s disease. J. Neurochem. 2014, 129, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Martorana, A.; Di Lorenzo, F.; Belli, L.; Sancesario, G.; Toniolo, S.; Sallustio, F.; Sancesario, G.M.; Koch, G. Cerebrospinal fluid Aβ42 levels: When physiological become pathological state. CNS Neurosci. Ther. 2015, 21, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Fedele, E.; Rivera, D.; Marengo, B.; Pronzato, M.A.; Ricciarelli, R. Amyloid β: Walking on the dark side of the moon. Mech. Ageing Dev. 2015, 152, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Goyal, R. Amyloid beta plaque: A culprit for neurodegeneration. Acta Neurol. Belg. 2016, 116, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed]
- Alkasir, R.; Li, J.; Li, X.; Jin, M.; Zhu, B. Human gut microbiota: The links with dementia development. Protein Cell 2017, 8, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Dudai, Y. Molecular bases of long-term memories: A question of persistence. Curr. Opin. Neurobiol. 2002, 12, 211–216. [Google Scholar] [CrossRef]
- Mrak, R.E.; Griffin, W.S. The role of activated astrocytes and of the neurotrophic cytokine S100B in the pathogenesis of Alzheimer’s disease. Neurobiol. Aging 2001, 22, 915–922. [Google Scholar] [CrossRef]
- Wilcock, D.M.; Griffin, W.S. Down’s syndrome, neuroinflammation, and Alzheimer neuropathogenesis. J. Neuroinflamm. 2013, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Donato, R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001, 33, 637–668. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. Testing times for the “amyloid cascade hypothesis”. Neurobiol. Aging 2002, 23, 1073–1074. [Google Scholar] [CrossRef]
- Selkoe, D.J.; American College of Physicians; American Physiological Society. Alzheimer disease: Mechanistic understanding predicts novel therapies. Ann. Intern. Med. 2004, 140, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Hayden, E.Y.; Teplow, D.B. Amyloid β-protein oligomers and Alzheimer’s disease. Alzheimers Res. Ther. 2013, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- van Dyck, C.H. Anti-amyloid-β monoclonal antibodies for Alzheimer’s disease: Pitfalls and promise. Biol. Psychiatry 2017. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.Y.; Fu, W.M. Drug candidates in clinical trials for Alzheimer’s disease. J. Biomed. Sci. 2017, 24, 47. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.A.; Vissel, B. Amyloid β: One of three danger-associated molecules that are secondary inducers of the proinflammatory cytokines that mediate Alzheimer’s disease. Br. J. Pharmacol. 2015, 172, 3714–3727. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.P.; Clark, I.A.; Vissel, B. Inconsistencies and controversies surrounding the amyloid hypothesis of Alzheimer’s disease. Acta Neuropathol. Commun. 2014, 2, 135. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.A.; Alleva, L.M.; Vissel, B. The roles of TNF in brain dysfunction and disease. Pharmacol. Ther. 2010, 128, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.A.; Vissel, B. Excess cerebral TNF causing glutamate excitotoxicity rationalizes treatment of neurodegenerative diseases and neurogenic pain by anti-TNF agents. J. Neuroinflamm. 2016, 13, 236. [Google Scholar] [CrossRef] [PubMed]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease and Down’s syndrome: Sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 1984, 122, 1131–1135. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W.; Quaranta, V.; Eanes, E.D. The amyloid deposits in Alzheimer’s disease: Their nature and pathogenesis. Appl. Pathol. 1984, 2, 357–369. [Google Scholar] [PubMed]
- Masters, C.L.; Multhaup, G.; Simms, G.; Pottgiesser, J.; Martins, R.N.; Beyreuther, K. Neuronal origin of a cerebral amyloid: Neurofibrillary tangles of Alzheimer’s disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J. 1985, 4, 2757–2763. [Google Scholar] [PubMed]
- Masters, C.L.; Simms, G.; Weinman, N.A.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249. [Google Scholar] [CrossRef] [PubMed]
- Narang, H.K. High-resolution electron microscopic analysis of the amyloid fibril in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 1980, 39, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Merz, P.A.; Wisniewski, H.M.; Somerville, R.A.; Bobin, S.A.; Masters, C.L.; Iqbal, K. Ultrastructural morphology of amyloid fibrils from neuritic and amyloid plaques. Acta Neuropathol. 1983, 60, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Shanmugam, A.; Bitan, G. Structure–function relationships of pre-fibrillar protein assemblies in Alzheimer’s disease and related disorders. Curr. Alzheimer Res. 2008, 5, 319–341. [Google Scholar] [CrossRef] [PubMed]
- Roychaudhuri, R.; Yang, M.; Hoshi, M.M.; Teplow, D.B. Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 2009, 284, 4749–4753. [Google Scholar] [CrossRef] [PubMed]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Bitan, G. Methods for studying and structure–function relationships of non-fibrillar protein assemblies in Alzheimer’s disease and related disorders. In Advances in Alzheimer Research; Lahiri, D.K., Ed.; Bentham Science Publishers: Sharjah, United Arab Emirates, 2014; Volume 2, pp. 291–374. ISBN 978-1-60-805853-2. [Google Scholar]
- Söderberg, L.; Bogdanovic, N.; Axelsson, B.; Winblad, B.; Näslund, J.; Tjernberg, L.O. Analysis of single Alzheimer solid plaque cores by laser capture microscopy and nanoelectrospray/tandem mass spectrometry. Biochemistry 2006, 45, 9849–9856. [Google Scholar] [CrossRef] [PubMed]
- Mitkevich, O.V.; Kochneva-Pervukhova, N.V.; Surina, E.R.; Benevolensky, S.V.; Kushnirov, V.V.; Ter-Avanesyan, M.D. DNA aptamers detecting generic amyloid epitopes. Prion 2012, 6, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Rüfenacht, P.; Güntert, A.; Bohrmann, B.; Ducret, A.; Döbeli, H. Quantification of the Aβ peptide in Alzheimer’s plaques by laser dissection microscopy combined with mass spectrometry. J. Mass Spectrom. 2005, 40, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Wirths, O.; Walter, S.; Kraus, I.; Klafki, H.W.; Stazi, M.; Oberstein, T.J.; Ghiso, J.; Wiltfang, J.; Bayer, T.A.; Weggen, S. N-truncated Aβ4−x peptides in sporadic Alzheimer’s disease cases and transgenic Alzheimer mouse models. Alzheimers Res. Ther. 2017, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Panchal, M.; Gaudin, M.; Lazar, A.N.; Salvati, E.; Rivals, I.; Ayciriex, S.; Dauphinot, L.; Dargere, D.; Auzeil, N.; Masserini, M.; et al. Ceramides and sphingomyelinases in senile plaques. Neurobiol. Dis. 2014, 65, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Panchal, M.; Loeper, J.; Cossec, J.C.; Perruchini, C.; Lazar, A.; Pompon, D.; Duyckaerts, C. Enrichment of cholesterol in microdissected Alzheimer’s disease senile plaques as assessed by mass spectrometry. J. Lipid Res. 2010, 51, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, A.T. Amyloid accomplices and enforcers. Protein Sci. 2005, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Cheng, D.; Wang, J.; Duong, D.M.; Losik, T.G.; Gearing, M.; Rees, H.D.; Lah, J.J.; Levey, A.I.; Peng, J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J. Biol. Chem. 2004, 279, 37061–37068. [Google Scholar] [CrossRef] [PubMed]
- Hadley, K.C.; Rakhit, R.; Guo, H.; Sun, Y.; Jonkman, J.E.; McLaurin, J.; Hazrati, L.N.; Emili, A.; Chakrabartty, A. Determining composition of micron-scale protein deposits in neurodegenerative disease by spatially targeted optical microproteomics. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.E.; Goyette, J.; Utter, V.; Rahimi, F.; Yang, Z.; Geczy, C.L.; Halliday, G.M. Inflammatory S100A9 and S100A12 proteins in Alzheimer’s disease. Neurobiol. Aging 2006, 27, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, A.; Hill, M.D.; Rahimi, F.; Warden, L.A.; Halliday, G.M.; Shepherd, C.E. Monocyte chemoattractant protein-1 plays a dominant role in the chronic inflammation observed in Alzheimer’s disease. Brain Pathol. 2009, 19, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Atwood, C.S.; Martins, R.N.; Smith, M.A.; Perry, G. Senile plaque composition and posttranslational modification of amyloid-β peptide and associated proteins. Peptides 2002, 23, 1343–1350. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Galvin, J.E.; Chiu, T.S.; Lee, V.M.; Masliah, E.; Trojanowski, J.Q. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998, 96, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, S.D.; Crino, P.B.; Hemby, S.E.; Weingarten, J.A.; Lee, V.M.; Eberwine, J.H.; Trojanowski, J.Q. Predominance of neuronal mRNAs in individual Alzheimer’s disease senile plaques. Ann. Neurol. 1999, 45, 174–181. [Google Scholar] [CrossRef]
- Marcinkiewicz, M. βAPP and furin mRNA concentrates in immature senile plaques in the brain of Alzheimer patients. J. Neuropathol. Exp. Neurol. 2002, 61, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Hirschfield, G.M.; Hawkins, P.N. Amyloidosis: New strategies for treatment. Int. J. Biochem. Cell Biol. 2003, 35, 1608–1613. [Google Scholar] [CrossRef]
- Drummond, E.; Nayak, S.; Faustin, A.; Pires, G.; Hickman, R.A.; Askenazi, M.; Cohen, M.; Haldiman, T.; Kim, C.; Han, X.; et al. Proteomic differences in amyloid plaques in rapidly progressive and sporadic Alzheimer’s disease. Acta Neuropathol. 2017, 133, 933–954. [Google Scholar] [CrossRef] [PubMed]
- Gerakis, Y.; Hetz, C. Emerging roles of ER stress in the etiology and pathogenesis of Alzheimer’s disease. FEBS J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gibas, K.J. The starving brain: Overfed meets undernourished in the pathology of mild cognitive impairment (MCI) and Alzheimer’s disease (AD). Neurochem. Int. 2017, 110, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Song, D.; Qing, H. Neural changes in Alzheimer’s disease from circuit to molecule: Perspective of optogenetics. Neurosci. Biobehav. Rev. 2017, 79, 110–118. [Google Scholar] [CrossRef] [PubMed]
- De Groot, N.S.; Burgas, M.T. Is membrane homeostasis the missing link between inflammation and neurodegenerative diseases? Cell. Mol. Life Sci. 2015, 72, 4795–4805. [Google Scholar] [CrossRef] [PubMed]
- Budimir, A. Metal ions, Alzheimer’s disease and chelation therapy. Acta Pharm. 2011, 61, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Petrou, A.L.; Terzidaki, A. A meta-analysis and review examining a possible role for oxidative stress and singlet oxygen in diverse diseases. Biochem. J. 2017, 474, 2713–2731. [Google Scholar] [CrossRef] [PubMed]
- Bagyinszky, E.; Giau, V.V.; Shim, K.; Suk, K.; An, S.S.A.; Kim, S. Role of inflammatory molecules in the Alzheimer’s disease progression and diagnosis. J. Neurol. Sci. 2017, 376, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Gutierrez, E.; Muñoz-Arenas, G.; Treviño, S.; Espinosa, B.; Chavez, R.; Rojas, K.; Flores, G.; Díaz, A.; Guevara, J. Alzheimer’s disease and metabolic syndrome: A link from oxidative stress and inflammation to neurodegeneration. Synapse 2017. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.E.; Ferreira, S.T. Crosstalk between endoplasmic reticulum stress and brain inflammation in Alzheimer’s disease. Neuropharmacology 2017. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Eddy, S.; Parma, D.; Gold, L. Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J. Mol. Biol. 1990, 213, 749–761. [Google Scholar] [CrossRef]
- Wrzesinski, J.; Jóźwiakowski, S.K. Structural basis for recognition of Co2+ by RNA aptamers. FEBS J. 2008, 275, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Niazi, J.H.; Chae, Y.J.; Ko, U.R.; Gu, M.B. Aptamers-in-liposomes for selective and multiplexed capture of small organic compounds. Macromol. Rapid Commun. 2011, 32, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bing, T.; Mei, H.; Fang, C.; Cao, Z.; Shangguan, D. Characterization and application of a DNA aptamer binding to l-tryptophan. Analyst 2011, 136, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Ohsawa, K.; Kasamatsu, T.; Shoji, A.; Sawai, H.; Ozaki, H. Screening of a glutamic acid-binding aptamer from arginine-modified DNA library. Nucleic Acids Symp. Ser. 2005, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Ames, T.D.; Breaker, R.R. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011, 8, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Darfeuille, F.; Hansen, J.B.; Orum, H.; Di Primo, C.; Toulmé, J.J. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004, 32, 3101–3107. [Google Scholar] [CrossRef] [PubMed]
- Lebars, I.; Richard, T.; Di Primo, C.; Toulmé, J.J. NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer. Nucleic Acids Res. 2007, 35, 6103–6114. [Google Scholar] [CrossRef] [PubMed]
- Lebars, I.; Richard, T.; Di Primo, C.; Toulmé, J.J. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells Mol. Dis. 2007, 38, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Sekkai, D.; Dausse, E.; Di Primo, C.; Darfeuille, F.; Boiziau, C.; Toulmé, J.J. In vitro selection of DNA aptamers against the HIV-1 TAR RNA hairpin. Antisense Nucleic Acid Drug Dev. 2002, 12, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P.; Phillips, T. Development of DNA aptamers to a foot-and-mouth disease peptide for competitive FRET-based detection. J. Biomol. Tech. 2008, 19, 109–115. [Google Scholar] [PubMed]
- Ashrafuzzaman, M. Aptamers as both drugs and drug-carriers. BioMed Res. Int. 2014, 2014, 697923. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, C.; Yin, J.; Li, B.; Zhou, M.; Dong, S.; Wang, E. Solid-state probe based electrochemical aptasensor for cocaine: A potentially convenient, sensitive, repeatable, and integrated sensing platform for drugs. Anal. Chem. 2010, 82, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, C.; Zhou, M.; Dong, S.; Wang, E. Microfluidic electrochemical aptameric assay integrated on-chip: A potentially convenient sensing platform for the amplified and multiplex analysis of small molecules. Anal. Chem. 2011, 83, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Osaki, T.; Sasaki, H.; Takinoue, M.; Yoshizawa, S.; Takeuchi, S. Rapid detection of a cocaine-binding aptamer using biological nanopores on a chip. J. Am. Chem. Soc. 2011, 133, 8474–8477. [Google Scholar] [CrossRef] [PubMed]
- Boese, B.J.; Breaker, R.R. In vitro selection and characterization of cellulose-binding DNA aptamers. Nucleic Acids Res. 2007, 35, 6378–6388. [Google Scholar] [CrossRef] [PubMed]
- Boese, B.J.; Corbino, K.; Breaker, R.R. In vitro selection and characterization of cellulose-binding RNA aptamers using isothermal amplification. Nucleosides Nucleotides Nucleic Acids 2008, 27, 949–966. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.P.; Kwon, I.K.; Sim, S.J. The strategy of signal amplification for ultrasensitive detection of hIgE based on aptamer-modified poly(di-acetylene) supramolecules. Biosens. Bioelectron. 2011, 26, 4823–4827. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.P.; Park, C.H.; Sim, S.J. Aptamer biosensors for label-free colorimetric detection of human IgE based on polydiacetylene (PDA) supramolecules. J. Nanosci. Nanotechnol. 2011, 11, 4269–4274. [Google Scholar] [CrossRef] [PubMed]
- Strahm, Y.; Flueckiger, A.; Billinger, M.; Meier, P.; Mettler, D.; Weisser, S.; Schaffner, T.; Hess, O. Endothelial-cell-binding aptamer for coating of intracoronary stents. J. Invasive Cardiol. 2010, 22, 481–487. [Google Scholar] [PubMed]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O., II; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K.W.; Giangrande, P.H. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 2009, 19, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Jyoti, A.; Vajpayee, P.; Singh, G.; Patel, C.B.; Gupta, K.C.; Shanker, R. Identification of environmental reservoirs of nontyphoidal salmonellosis: Aptamer-assisted bioconcentration and subsequent detection of Salmonella Typhimurium by quantitative polymerase chain reaction. Environ. Sci. Technol. 2011, 45, 8996–9002. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.Q.; Ren, Q.; Wei, H.P.; Chen, Z.; Deng, J.Y.; Zhang, Z.P.; Zhang, X.E. Quantum dot–aptamer nanoprobes for recognizing and labeling influenza A virus particles. Nanoscale 2011, 3, 2454–2457. [Google Scholar] [CrossRef] [PubMed]
- Ellenbecker, M.; Sears, L.; Li, P.; Lanchy, J.M.; Lodmell, J.S. Characterization of RNA aptamers directed against the nucleocapsid protein of Rift Valley fever virus. Antiviral Res. 2012, 93, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.H. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS ONE 2011, 6, e27862. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, S.; Yoon, H.; Kim, K.B.; Kalme, S.S.; Oh, S.; Song, C.S.; Kim, D.E. Selection of an antiviral RNA aptamer against hemagglutinin of the subtype H5 avian influenza virus. Nucleic Acid Ther. 2011, 21, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Diafa, S.; Hollenstein, M. Generation of aptamers with an expanded chemical repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M. DNA catalysis: The chemical repertoire of DNAzymes. Molecules 2015, 20, 20777–20804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.Y.; Acquah, C.; Sidhu, A.; Ongkudon, C.M.; Yon, L.S.; Danquah, M.K. SELEX modifications and bioanalytical techniques for aptamer–target binding characterization. Crit. Rev. Anal. Chem. 2016, 46, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wen, W.; Wu, A.; Niu, L. Chemically modified, α-amino-3-hydroxy-5-methyl-4-isoxazole (AMPA) receptor RNA aptamers designed for in vivo use. ACS Chem. Neurosci. 2017, 8, 2437–2445. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Rosenthal, M.; Siegl, J.; Ewers, J.; Mayer, G. Customised nucleic acid libraries for enhanced aptamer selection and performance. Curr. Opin. Biotechnol. 2017, 48, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Jenison, R.D.; Gill, S.C.; Pardi, A.; Polisky, B. High-resolution molecular discrimination by RNA. Science 1994, 263, 1425–1429. [Google Scholar] [CrossRef] [PubMed]
- Shoji, A.; Kuwahara, M.; Ozaki, H.; Sawai, H. Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J. Am. Chem. Soc. 2007, 129, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, T. The selection of aptamers specific for membrane molecular targets. Cell. Mol. Biol. Lett. 2011, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Macedo, B.; Cordeiro, Y. Unraveling prion protein interactions with aptamers and other PrP-binding nucleic acids. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Ylera, F.; Lurz, R.; Erdmann, V.A.; Fürste, J.P. Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem. Biophys. Res. Commun. 2002, 290, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Tada, K.; Mihara, H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 2009, 5, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Farrar, C.T.; William, C.M.; Hudry, E.; Hashimoto, T.; Hyman, B.T. RNA aptamer probes as optical imaging agents for the detection of amyloid plaques. PLoS ONE 2014, 9, e89901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, S.; Proske, D.; Neumann, M.; Groschup, M.H.; Kretzschmar, H.A.; Famulok, M.; Winnacker, E.L. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 1997, 71, 8790–8797. [Google Scholar] [PubMed]
- Rhie, A.; Kirby, L.; Sayer, N.; Wellesley, R.; Disterer, P.; Sylvester, I.; Gill, A.; Hope, J.; James, W.; Tahiri-Alaoui, A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 2003, 278, 39697–39705. [Google Scholar] [CrossRef] [PubMed]
- Takemura, K.; Wang, P.; Vorberg, I.; Surewicz, W.; Priola, S.A.; Kanthasamy, A.; Pottathil, R.; Chen, S.G.; Sreevatsan, S. DNA aptamers that bind to PrPC and not PrPSc show sequence and structure specificity. Exp. Biol. Med. 2006, 231, 204–214. [Google Scholar] [CrossRef]
- Bibby, D.F.; Gill, A.C.; Kirby, L.; Farquhar, C.F.; Bruce, M.E.; Garson, J.A. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J. Virol. Methods 2008, 151, 107–115. [Google Scholar] [CrossRef] [PubMed]
- King, D.J.; Safar, J.G.; Legname, G.; Prusiner, S.B. Thioaptamer interactions with prion proteins: Sequence-specific and non-specific binding sites. J. Mol. Biol. 2007, 369, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Gilch, S.; Wopfner, F.; Schätzl, H.M.; Winnacker, E.L.; Famulok, M. Prion-protein-specific aptamer reduces PrPSc formation. ChemBioChem 2002, 3, 717–725. [Google Scholar] [CrossRef]
- Murakami, K.; Nishikawa, F.; Noda, K.; Yokoyama, T.; Nishikawa, S. Anti-bovine prion protein RNA aptamer containing tandem GGA repeat interacts both with recombinant bovine prion protein and its β isoform with high affinity. Prion 2008, 2, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Lührs, T.; Zahn, R.; Wüthrich, K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J. Mol. Biol. 2006, 357, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.; Mantle, B.J.; Morten, I.J.; Tennent, G.A.; Radford, S.E.; Stockley, P.G. Production and characterization of RNA aptamers specific for amyloid fibril epitopes. J. Biol. Chem. 2007, 282, 34500–34509. [Google Scholar] [CrossRef] [PubMed]
- Tsukakoshi, K.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 2012, 84, 5542–5547. [Google Scholar] [CrossRef] [PubMed]
- Tsukakoshi, K.; Harada, R.; Sode, K.; Ikebukuro, K. Screening of DNA aptamer which binds to α-synuclein. Biotechnol. Lett. 2010, 32, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Burdick, D.; Soreghan, B.; Kwon, M.; Kosmoski, J.; Knauer, M.; Henschen, A.; Yates, J.; Cotman, C.; Glabe, C. Assembly and aggregation properties of synthetic Alzheimer’s A4/β amyloid peptide analogs. J. Biol. Chem. 1992, 267, 546–554. [Google Scholar] [PubMed]
- Stine, W.B.; Jungbauer, L.; Yu, C.; LaDu, M.J. Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 2011, 670, 13–32. [Google Scholar] [CrossRef] [PubMed]
- Mahood, R.A. Selection of RNA Aptamers and Their Recognition of Amyloid Assemblies. Ph.D. Thesis, The University of Leeds, Leeds, UK, 2015. [Google Scholar]
- Teplow, D.B. Preparation of amyloid β-protein for structural and functional studies. Methods Enzymol. 2006, 413, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Miller, D.L.; Sapienza, V.J.; Chen, C.M.J.; Bai, C.; Grundke-Iqbal, I.; Currie, J.R.; Wisniewski, H.M. Production and characterization of monoclonal antibodies reactive to synthetic cerebrovascular amyloid peptide. Neurosci. Res. Commun. 1988, 2, 121–130. [Google Scholar]
- Kim, K.S.; Wen, G.Y.; Bancher, C.; Chen, C.M.J.; Sapienza, V.J.; Hong, H.; Wisniewski, H.M. Detection and quantitation of amyloid β-peptide with two monoclonal antibodies. Neurosci. Res. Commun. 1990, 7, 113–122. [Google Scholar]
- Pirttilä, T.; Kim, K.S.; Mehta, P.D.; Frey, H.; Wisniewski, H.M. Soluble amyloid β-protein in the cerebrospinal fluid from patients with Alzheimer’s disease, vascular dementia and controls. J. Neurol. Sci. 1994, 127, 90–95. [Google Scholar] [CrossRef]
- Necula, M.; Kayed, R.; Milton, S.; Glabe, C.G. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007, 282, 10311–10324. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.; Aravind, A.; Brahatheeswaran, D.; Fukuda, T.; Nagaoka, Y.; Hasumura, T.; Iwai, S.; Morimoto, H.; Yoshida, Y.; Maekawa, T.; et al. Amyloid-binding aptamer conjugated curcumin—PLGA nanoparticle for potential use in Alzheimer’s disease. BioNanoScience 2012, 2, 83–93. [Google Scholar] [CrossRef]
- Chakravarthy, M.; Chen, S.; Dodd, P.R.; Veedu, R.N. Nucleic acid-based theranostics for tackling Alzheimer’s disease. Theranostics 2017, 7, 3933–3947. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Li, H.; Sinha, S.; Bitan, G. Modulators of Amyloid β-Protein (Aβ) Self-Assembly. In Developing Therapeutics for Alzheimer’s Diseas; Wolfe, M.S., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 97–191. ISBN 978-0-12-802173-6. [Google Scholar]
- Baell, J.; Walters, M.A. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The essential medicinal chemistry of curcumin. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar] [CrossRef] [PubMed]
- Bahadori, F.; Demiray, M. A realistic view on “the essential medicinal chemistry of curcumin”. ACS Med. Chem. Lett. 2017, 8, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Murakami, K.; Summers, J.L.; Chen, C.H.B.; Bitan, G. RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS ONE 2009, 4, e7694. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Lomakin, A.; Teplow, D.B. Amyloid β-protein oligomerization: Prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J. Biol. Chem. 2001, 276, 35176–35184. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Maiti, P.; Bitan, G. Photo-induced cross-linking of unmodified proteins (PICUP) applied to amyloidogenic peptides. J. Vis. Exp. 2009. [Google Scholar] [CrossRef] [PubMed]
- Rosensweig, C.; Ono, K.; Murakami, K.; Lowenstein, D.K.; Bitan, G.; Teplow, D.B. Preparation of stable amyloid β-protein oligomers of defined assembly order. Methods Mol. Biol. 2012, 849, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Fradinger, E.A.; Spring, S.M.; Teplow, D.B. Neurotoxic protein oligomers—What you see is not always what you get. Amyloid 2005, 12, 88–95. [Google Scholar] [CrossRef] [PubMed]
- LeVine, H., 3rd. Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999, 309, 274–284. [Google Scholar] [PubMed]
- Li, H.; Rahimi, F.; Sinha, S.; Maiti, P.; Bitan, G.; Murakami, K. Amyloids and Protein Aggregation—Analytical Methods. In Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2009; ISBN 9780470027318. [Google Scholar]
- Rahimi, F.; Bitan, G. Selection of aptamers for amyloid β-protein, the causative agent of Alzheimer’s disease. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [PubMed]
- Jankowsky, J.L.; Fadale, D.J.; Anderson, J.; Xu, G.M.; Gonzales, V.; Jenkins, N.A.; Copeland, N.G.; Lee, M.K.; Younkin, L.H.; Wagner, S.L.; et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: Evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 2004, 13, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Klunk, W.E.; Bacskai, B.J.; Mathis, C.A.; Kajdasz, S.T.; McLellan, M.E.; Frosch, M.P.; Debnath, M.L.; Holt, D.P.; Wang, Y.; Hyman, B.T. Imaging Aβ plaques in living transgenic mice with multiphoton microscopy and methoxy-X04, a systemically administered Congo red derivative. J. Neuropathol. Exp. Neurol. 2002, 61, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Koffie, R.M.; Meyer-Luehmann, M.; Hashimoto, T.; Adams, K.W.; Mielke, M.L.; Garcia-Alloza, M.; Micheva, K.D.; Smith, S.J.; Kim, M.L.; Lee, V.M.; et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. USA 2009, 106, 4012–4017. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.B.; Leng, L.Z.; Zhang, B.; Kwong, L.; Trojanowski, J.Q.; Abel, T.; Lee, V.M. Targeting amyloid-β peptide (Aβ) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Aβ precursor protein (APP) transgenic mice. J. Biol. Chem. 2006, 281, 4292–4299. [Google Scholar] [CrossRef] [PubMed]
- Gudiksen, K.L.; Gitlin, I.; Whitesides, G.M. Differentiation of proteins based on characteristic patterns of association and denaturation in solutions of SDS. Proc. Natl. Acad. Sci. USA 2006, 103, 7968–7972. [Google Scholar] [CrossRef] [PubMed]
- Leffers, K.W.; Schell, J.; Jansen, K.; Lucassen, R.; Kaimann, T.; Nagel-Steger, L.; Tatzelt, J.; Riesner, D. The structural transition of the prion protein into its pathogenic conformation is induced by unmasking hydrophobic sites. J. Mol. Biol. 2004, 344, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Kawooya, J.K.; Emmons, T.L.; Gonzalez-DeWhitt, P.A.; Camp, M.C.; D’Andrea, S.C. Electrophoretic mobility of Alzheimer’s amyloid-β peptides in urea-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 2003, 323, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Montserret, R.; McLeish, M.J.; Böckmann, A.; Geourjon, C.; Penin, F. Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding. Biochemistry 2000, 39, 8362–8373. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Hasegawa, K.; Yamaguchi, I.; Tsutsumi, S.; Kardos, J.; Goto, Y.; Gejyo, F.; Naiki, H. Low concentrations of sodium dodecyl sulfate induce the extension of β2-microglobulin-related amyloid fibrils at a neutral pH. Biochemistry 2004, 43, 11075–11082. [Google Scholar] [CrossRef] [PubMed]
- Rangachari, V.; Moore, B.D.; Reed, D.K.; Sonoda, L.K.; Bridges, A.W.; Conboy, E.; Hartigan, D.; Rosenberry, T.L. Amyloid-β(1–42) rapidly forms protofibrils and oligomers by distinct pathways in low concentrations of sodium dodecylsulfate. Biochemistry 2007, 46, 12451–12462. [Google Scholar] [CrossRef] [PubMed]
- Rangachari, V.; Reed, D.K.; Moore, B.D.; Rosenberry, T.L. Secondary structure and interfacial aggregation of amyloid-β(1–40) on sodium dodecyl sulfate micelles. Biochemistry 2006, 45, 8639–8648. [Google Scholar] [CrossRef] [PubMed]
- Piening, N.; Weber, P.; Högen, T.; Beekes, M.; Kretzschmar, H.; Giese, A. Photo-induced crosslinking of prion protein oligomers and prions. Amyloid 2006, 13, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Moussa, C.E.; Wersinger, C.; Rusnak, M.; Tomita, Y.; Sidhu, A. Abnormal migration of human wild-type α-synuclein upon gel electrophoresis. Neurosci. Lett. 2004, 371, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Podlisny, M.B.; Ostaszewski, B.L.; Squazzo, S.L.; Koo, E.H.; Rydell, R.E.; Teplow, D.B.; Selkoe, D.J. Aggregation of secreted amyloid β-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J. Biol. Chem. 1995, 270, 9564–9570. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.M.; Hartley, D.M.; Kusumoto, Y.; Fezoui, Y.; Condron, M.M.; Lomakin, A.; Benedek, G.B.; Selkoe, D.J.; Teplow, D.B. Amyloid β-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 1999, 274, 25945–25952. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Takio, K.; Ogawara, M.; Selkoe, D.J. Mass spectrometry of purified amyloid β protein in Alzheimer’s disease. J. Biol. Chem. 1992, 267, 17082–17086. [Google Scholar] [PubMed]
- Yu, L.; Edalji, R.; Harlan, J.E.; Holzman, T.F.; Lopez, A.P.; Labkovsky, B.; Hillen, H.; Barghorn, S.; Ebert, U.; Richardson, P.L.; et al. Structural characterization of a soluble amyloid β-peptide oligomer. Biochemistry 2009, 48, 1870–1877. [Google Scholar] [CrossRef] [PubMed]
- Barghorn, S.; Nimmrich, V.; Striebinger, A.; Krantz, C.; Keller, P.; Janson, B.; Bahr, M.; Schmidt, M.; Bitner, R.S.; Harlan, J.; et al. Globular amyloid β-peptide1–42 oligomer—A homogenous and stable neuropathological protein in Alzheimer’s disease. J. Neurochem. 2005, 95, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Kirkitadze, M.D.; Lomakin, A.; Vollers, S.S.; Benedek, G.B.; Teplow, D.B. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA 2003, 100, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Tarus, B.; Vollers, S.S.; Lashuel, H.A.; Condron, M.M.; Straub, J.E.; Teplow, D.B. A molecular switch in amyloid assembly: Met35 and amyloid β-protein oligomerization. J. Am. Chem. Soc. 2003, 125, 15359–15365. [Google Scholar] [CrossRef] [PubMed]
- Hepler, R.W.; Grimm, K.M.; Nahas, D.D.; Breese, R.; Dodson, E.C.; Acton, P.; Keller, P.M.; Yeager, M.; Wang, H.; Shughrue, P.; et al. Solution state characterization of amyloid β-derived diffusible ligands. Biochemistry 2006, 45, 15157–15167. [Google Scholar] [CrossRef] [PubMed]
- O’Nuallain, B.; Freir, D.B.; Nicoll, A.J.; Risse, E.; Ferguson, N.; Herron, C.E.; Collinge, J.; Walsh, D.M. Amyloid β-protein dimers rapidly form stable synaptotoxic protofibrils. J. Neurosci. 2010, 30, 14411–14419. [Google Scholar] [CrossRef] [PubMed]
- Watt, A.D.; Perez, K.A.; Rembach, A.; Sherrat, N.A.; Hung, L.W.; Johanssen, T.; McLean, C.A.; Kok, W.M.; Hutton, C.A.; Fodero-Tavoletti, M.; et al. Oligomers, fact or artefact? SDS-PAGE induces dimerization of β-amyloid in human brain samples. Acta Neuropathol. 2013, 125, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Pujol-Pina, R.; Vilaprinyó-Pascual, S.; Mazzucato, R.; Arcella, A.; Vilaseca, M.; Orozco, M.; Carulla, N. SDS-PAGE analysis of Aβ oligomers is disserving research into Alzheimer s disease: Appealing for ESI-IM-MS. Sci. Rep. 2015, 5, 14809. [Google Scholar] [CrossRef] [PubMed]
- Bitan, G.; Vollers, S.S.; Teplow, D.B. Elucidation of primary structure elements controlling early amyloid β-protein oligomerization. J. Biol. Chem. 2003, 278, 34882–34889. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, K.N.; Manelli, A.M.; Stine, W.B., Jr.; Baker, L.K.; Krafft, G.A.; LaDu, M.J. Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 2002, 277, 32046–32053. [Google Scholar] [CrossRef] [PubMed]
- Vasilevko, V.; Pop, V.; Kim, H.J.; Saing, T.; Glabe, C.C.; Milton, S.; Barrett, E.G.; Cotman, C.W.; Cribbs, D.H.; Head, E. Linear and conformation specific antibodies in aged beagles after prolonged vaccination with aggregated Abeta. Neurobiol. Dis. 2010, 39, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Head, E.; Pop, V.; Vasilevko, V.; Hill, M.; Saing, T.; Sarsoza, F.; Nistor, M.; Christie, L.A.; Milton, S.; Glabe, C.; et al. A two-year study with fibrillar β-amyloid (Aβ) immunization in aged canines: Effects on cognitive function and brain Aβ. J. Neurosci. 2008, 28, 3555–3566. [Google Scholar] [CrossRef] [PubMed]
- Galzitskaya, O.V. Repeats are one of the main characteristics of RNA-binding proteins with prion-like domains. Mol. Biosyst. 2015, 11, 2210–2218. [Google Scholar] [CrossRef] [PubMed]
- Calamai, M.; Kumita, J.R.; Mifsud, J.; Parrini, C.; Ramazzotti, M.; Ramponi, G.; Taddei, N.; Chiti, F.; Dobson, C.M. Nature and significance of the interactions between amyloid fibrils and biological polyelectrolytes. Biochemistry 2006, 45, 12806–12815. [Google Scholar] [CrossRef] [PubMed]
- Maury, C.P. The emerging concept of functional amyloid. J. Intern. Med. 2009, 265, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Sharon, R.; Bar-Joseph, I.; Frosch, M.P.; Walsh, D.M.; Hamilton, J.A.; Selkoe, D.J. The formation of highly soluble oligomers of α-synuclein is regulated by fatty acids and enhanced in Parkinson’s disease. Neuron 2003, 37, 583–595. [Google Scholar] [CrossRef]
- Lesné, S.; Koh, M.T.; Kotilinek, L.; Kayed, R.; Glabe, C.G.; Yang, A.; Gallagher, M.; Ashe, K.H. A specific amyloid-β protein assembly in the brain impairs memory. Nature 2006, 440, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Klucken, J.; Ingelsson, M.; Shin, Y.; Irizarry, M.C.; Hedley-Whyte, E.T.; Frosch, M.; Growdon, J.; McLean, P.; Hyman, B.T. Clinical and biochemical correlates of insoluble α-synuclein in dementia with Lewy bodies. Acta Neuropathol. 2006, 111, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Bloodgood, B.L.; Townsend, M.; Walsh, D.M.; Selkoe, D.J.; Sabatini, B.L. Natural oligomers of the Alzheimer amyloid-β protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J. Neurosci. 2007, 27, 2866–2875. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Li, S.; Mehta, T.H.; Garcia-Munoz, A.; Shepardson, N.E.; Smith, I.; Brett, F.M.; Farrell, M.A.; Rowan, M.J.; Lemere, C.A.; et al. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008, 14, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Paleologou, K.E.; Kragh, C.L.; Mann, D.M.; Salem, S.A.; Al-Shami, R.; Allsop, D.; Hassan, A.H.; Jensen, P.H.; El-Agnaf, O.M. Detection of elevated levels of soluble α-synuclein oligomers in post-mortem brain extracts from patients with dementia with Lewy bodies. Brain 2009, 132, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Head, E.; Pop, V.; Sarsoza, F.; Kayed, R.; Beckett, T.L.; Studzinski, C.M.; Tomic, J.L.; Glabe, C.G.; Murphy, M.P. Amyloid-β peptide and oligomers in the brain and cerebrospinal fluid of aged canines. J. Alzheimers Dis. 2010, 20, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Condron, M.M.; Teplow, D.B. Structure–neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl. Acad. Sci. USA 2009, 106, 14745–14750. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Yu, S.; Zheng, Y.; Zheng, Y.; Yang, H.; Zhang, J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017, 74, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Gasse, C.; Hollenstein, M. Nucleic acid aptamers: Emerging applications in medical imaging, nanotechnology, neurosciences, and drug delivery. Int. J. Mol. Sci. 2017, 18, 2430. [Google Scholar] [CrossRef] [PubMed]
- Sriramoju, B.; Kanwar, R.; Veedu, R.N.; Kanwar, J.R. Aptamer-targeted oligonucleotide theranostics: A smarter approach for brain delivery and the treatment of neurological diseases. Curr. Top. Med. Chem. 2015, 15, 1115–1124. [Google Scholar] [CrossRef] [PubMed]

| Aptamer Type | Target | SELEX Method | Aptamer Reactivity | Reference |
|---|---|---|---|---|
| RNA, β aptamers, e.g., β55 | Synthetic Aβ40 with an engineered N-terminal cysteine | Chromatographic separation using Sepharose 6B matrix carrying the target | No interaction with monomeric, soluble Aβ40, but reactive with Aβ40 fibrils | [125] |
| RNA aptamers E1, E2, N1, G2 etc. | Aβ40 conjugated to gold nanoparticles as a model of Aβ oligomers | RNA pool was exposed to target, separation was by centrifugation, and three different elution strategies used | Aβ40 oligomer model and apparently monomeric Aβ40 | [126] |
| RNA aptamers, KM and previously reported β aptamers | PICUP-generated and purified trimeric Aβ40, and a PICUP-generated mixture of low-molecular-weight Aβ40 oligomers | Filter-binding assay used for separation | Aβ fibrils and fibrils of other exemplary amyloidogenic proteins | [153,160] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahimi, F. Aptamers Selected for Recognizing Amyloid β-Protein—A Case for Cautious Optimism. Int. J. Mol. Sci. 2018, 19, 668. https://doi.org/10.3390/ijms19030668
Rahimi F. Aptamers Selected for Recognizing Amyloid β-Protein—A Case for Cautious Optimism. International Journal of Molecular Sciences. 2018; 19(3):668. https://doi.org/10.3390/ijms19030668
Chicago/Turabian StyleRahimi, Farid. 2018. "Aptamers Selected for Recognizing Amyloid β-Protein—A Case for Cautious Optimism" International Journal of Molecular Sciences 19, no. 3: 668. https://doi.org/10.3390/ijms19030668
APA StyleRahimi, F. (2018). Aptamers Selected for Recognizing Amyloid β-Protein—A Case for Cautious Optimism. International Journal of Molecular Sciences, 19(3), 668. https://doi.org/10.3390/ijms19030668