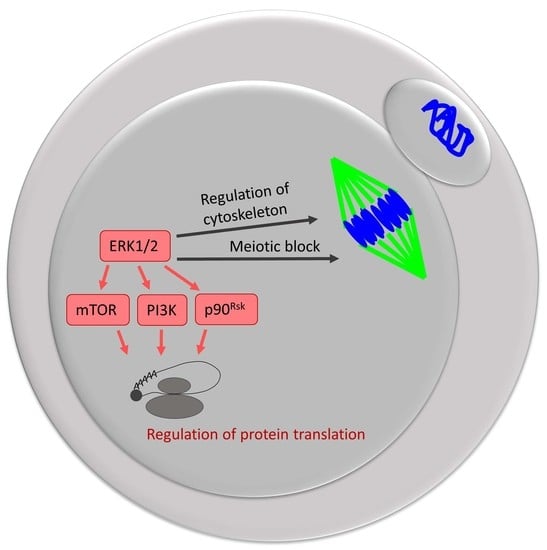
Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis
Abstract
:1. Introduction
2. MAP Kinases
3. Role of MAPK in Regulation of Meiosis
4. Translational Regulation at 5′ End of mRNA (Cap-Dependent Initiation of Protein Translation)
5. Translational Regulation at the 3′ End of mRNA
6. eIF4E Activity in Oocytes
7. Role of ERK1/2 in eIF4E Activation in Meiotic Cells
8. Cytoplasmic Polyadenylation of mRNA in Oocytes
9. A Role of ERK1/2 in mRNA Polyadenylation in Oocytes
10. Cross-Talk between ERK1/2 and PI3K/mTOR Pathways
11. Perspectives
Acknowledgments
Conflicts of Interest
Abbreviations
| ERK1/2 | The extracellular regulated kinases 1 and 2 |
| MAPK | Mitogen activated protein kinase |
| MAPKK | MAP kinases kinase |
| MAPKKK | MAP kinase |
| RSK | p90 ribosomal S6 kinase |
| MNKs | MAPK-interacting kinases MNK1 and MNK2 |
| S6K | p70 S6 kinase |
| CSF | Cytostatic factor |
| mTOR | mammalian target of rapamycin |
| eIFs | Eukaryotic initiation factors |
| eIF4F | Eukaryotic translation initiation complex 4F |
| eIF4E | Eukaryotic translation initiation factor 4E |
| eIF4B | Eukaryotic translation initiation factor 4B |
| eIF4A | Eukaryotic translation initiation factor 4A |
| eIF4G1 | Eukaryotic initiation factor 4G1 |
| 4E-BPs | EIF4E-binding proteins |
| 4E-BP1 | EIF4E |
| CPE | Cytoplasmic polyadenylation element |
| CPEB | CPE-binding protein |
| CPEB1 | CPE-binding protein 1 |
| PI3K | Phosphatidylinositol 3-kinase |
| PKB | Protein kinase B |
| MOS | Oocyte maturation factor Mos |
| MISS | MAPK interacting and spindle stabilizing |
| DOC1R | Deleted in oral cancer 1 related |
| APC/C | Anaphase-promoting complex/cyclosome |
| MPF | Maturation promoting factor |
| m7G | 7-methylguanylate |
| MI | Meiosis I |
| MII | Meiosis II |
| PAP | Poly(A) polymerase |
| CPSF | Cytoplasmic polyadenylation specific factor |
| PP2A | Protein phosphatase 2A |
| Ras | Rat sarcoma virus oncogene |
| Raf | MEK kinase Raf |
| KSR | Kinase suppressor of Ras |
| MP1 | MEK scaffolding protein 1 |
| TSC 2 | Tuberous sclerosis complex 2 |
| RAPTOR | Regulatory associated protein of mTOR |
References
- Tanaka, M.; Kihara, M.; Hennebold, J.D.; Eppig, J.J.; Viveiros, M.M.; Emery, B.R.; Carrell, D.T.; Kirkman, N.J.; Meczekalski, B.; Zhou, J.; et al. H1FOO is coupled to the initiation of oocytic growth. Biol. Reprod. 2005, 72, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, Z.; Kageyama, S.I.; Aoki, F. Degradation of maternal mRNA in mouse embryos: Selective degradation of specific mRNAs after fertilization. Mol. Reprod. Dev. 2005, 72, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Melton, C.; Suh, N.; Oh, J.S.; Horner, K.; Xie, F.; Sette, C.; Blelloch, R.; Conti, M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011, 25, 755–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheets, M.D.; Fox, C.A.; Hunt, T.; Vande Woude, G.; Wickens, M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994, 8, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Lasko, P. Translational control in oocyte development. Cold Spring Harb. Perspect. Biol. 2011, 3, a002758. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J. Mouse Development; Springer: Berlin/Heidelberg, Germany, 2012; Volume 55, pp. 1–21. [Google Scholar] [CrossRef]
- Hamatani, T.; Daikoku, T.; Wang, H.; Matsumoto, H.; Carter, M.G.; Ko, M.S.H.; Dey, S.K. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. USA 2004, 101, 10326–10331. [Google Scholar] [CrossRef] [PubMed]
- Mehlmann, L.M. Stops and starts in mammalian oocytes: Recent advances in understanding the regulation of meiotic arrest and oocyte maturation. Reproduction 2005, 130, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Piqué, M.; López, J.M.; Foissac, S.; Guigó, R.; Méndez, R. A Combinatorial Code for CPE-Mediated Translational Control. Cell 2008, 132, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Gosden, R.; Lee, B. Portrait of an oocyte: Our obscure origin. J. Clin. Investig. 2010, 120, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Torcia, S.; Xie, F.; Lin, C.J.; Cakmak, H.; Franciosi, F.; Horner, K.; Onodera, C.; Song, J.S.; Cedars, M.I.; et al. Somatic cells regulate maternal mRNA translation and developmental competence of mouse oocytes. Nat. Cell Biol. 2013, 15, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Piccioni, F.; Zappavigna, V.; Verrotti, A.C. Translational regulation during oogenesis and early development: The cap-poly(A) tail relationship. C. R. Biol. 2005, 328, 863–881. [Google Scholar] [CrossRef] [PubMed]
- Barkoff, A.F.; Dickson, K.S.; Gray, N.K.; Wickens, M. Translational control of cyclin B1 mRNA during meiotic maturation: Coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 2000, 220, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Gorbsky, G.J. The spindle checkpoint and chromosome segregation in meiosis. FEBS J. 2015, 282, 2471–2487. [Google Scholar] [CrossRef] [PubMed]
- Schultz, G.A.; Clough, J.R.; Johnson, M.H. Presence of cap structures in the messenger RNA of mouse eggs. Development 1980, 56, 139–156. [Google Scholar]
- Radford, H.E.; Meijer, H.A.; de Moor, C.H. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2008, 1779, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Brook, M.; Smith, J.W.S.; Gray, N.K. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 2009, 137, 595–617. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, J.M.; Avruch, J. Mammalian MAPK Signal Transduction Pathways Activated by Stress and Inflammation: A 10-Year Update. Physiol. Rev. 2012, 92, 689–737. [Google Scholar] [CrossRef] [PubMed]
- Peti, W.; Page, R. Molecular basis of MAP kinase regulation. Protein Sci. 2013, 22, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shepherd, E.G.; Nelin, L.D. MAPK phosphatases—Regulating the immune response. Nat. Rev. Immunol. 2007, 7, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.S.C.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.L. Defining MAPK interactomes. ACS Chem. Biol. 2011, 6, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Pimienta, G.; Pascual, J. Canonical and alternative MAPK signalling. Cell Cycle 2007, 6, 2628–2632. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cobb, M.H. Regulation of stress-responsive mitogen-activated protein (MAP) kinase pathways by TAO2. J. Biol. Chem. 2001, 276, 16070–16075. [Google Scholar] [CrossRef] [PubMed]
- Pearson, G.; Robinson, F.; Beers Gibson, T.; Xu, B.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-Activated Protein (MAP) Kinase Pathways: Regulation and Physiological Functions. Endocr. Rev. 2001, 22, 153–183. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.M.; Keyse, S.M. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007, 26, 3203–3213. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.L.; Nakamura, K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Shaul, Y.D.; Seger, R. The MEK/ERK cascade: From signalling specificity to diverse functions. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2007, 1773, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Knight, T.; Irving, J.A.E. Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and Its Therapeutic Targeting. Front. Oncol. 2014, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef] [PubMed]
- Gaestel, M. MAPK-Activated Protein Kinases (MKs): Novel Insights and Challenges. Front. Cell Dev. Biol. 2016, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Anjum, R.; Blenis, J. The RSK family of kinases: Emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008, 9, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Waskiewicz, A.J.; Flynn, A.; Proud, C.G.; Cooper, J.A. Mitogen-activated protein kinases activate the serine/threonine kinaseses Mnk1 and Mnk2. EMBO J. 1997, 16, 1909–1920. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Topisirovic, I. Regulation of mRNA translation by signalling pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Buxade, M.; Morrice, N.; Krebs, D.L.; Proud, C.G. The PSF·p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor α. J. Biol. Chem. 2008, 283, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Scheper, G.C.; Morrice, N.A.; Kleijn, M.; Proud, C.G. The Mitogen-Activated Protein Kinase Signal-Integrating Kinase Mnk2 Is a Eukaryotic Initiation Factor 4E Kinase with High Levels of Basal Activity in Mammalian Cells The Mitogen-Activated Protein Kinase Signal-Integrating Kinase Mnk2 Is a Eukaryotic Initi. Mol. Cell. Biol. 2001, 21, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.Z.; Harms, U.; Klostermeier, D. eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region. RNA Biol. 2017, 14, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, B.; Ekim, B.; Fingar, D.C. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 2012, 441, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Inition in Eukaryotes: Mechanisms and Biological Targrts. Cell 2013, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Masuyama, N.; Dell, K.; Shirakabe, K.; Nishida, E. Initiation of Xenopus oocyte maturation by activation of the mitogen-activated protein kinase cascade. J Biol. Chem. 1995, 270, 25898–25904. [Google Scholar] [CrossRef] [PubMed]
- Kubelka, M.; Anger, M.; Kalous, J.; Schultz, R.M.; Motlík, J. Chromosome condensation in pig oocytes: Lack of a requirement for either cdc2 kinase or MAP kinase activity. Mol. Reprod. Dev. 2002, 63, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Kalous, J.; Kubelka, M.; Motlík, J. The effect of PD98059 on MAPK regulation in cumulus-enclosed and cumulus-free mouse oocytes. Zygote 2003, 11, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-Y.; Sun, Q.-Y. Involvement of Mitogen-Activated Protein Kinase Cascade During Oocyte Maturation and Fertilization in Mammals1. Biol. Reprod. 2004, 70, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Liu, X.M.; Ji, S.Y.; Sha, Q.Q.; Zhang, J.; Fan, H.Y. ERK1/2 Activities Are Dispensable for Oocyte Growth but Are Required for Meiotic Maturation and Pronuclear Formation in Mouse. J. Genet. Genom. 2015, 42, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Verlhac, M.H.; Kubiak, J.Z.; Weber, M.; Géraud, G.; Colledge, W.H.; Evans, M.J.; Maro, B. Mos is required for MAP kinase activation and is involved in microtubule organization during meiotic maturation in the mouse. Development 1996, 122, 815–822. [Google Scholar] [PubMed]
- Paronetto, M.P.; Giorda, E.; Carsetti, R.; Rossi, P.; Geremia, R.; Sette, C. Functional interaction between p90Rsk2 and Emi1 contributes to the metaphase arrest of mouse oocytes. EMBO J. 2004, 23, 4649–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, C.; Emilie Terret, M.; Djiane, A.; Rassinier, P.; Maro, B.; Verlhac, M.H. Meiotic spindle stability depends on MAPK-interacting and spindle-stabilizing protein (MISS), a new MAPK substrate. J. Cell Biol. 2002, 157, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Terret, M.E. DOC1R: A MAP kinase substrate that control microtubule organization of metaphase II mouse oocytes. Development 2003, 130, 5169–5177. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A. Cytostatic factor: An activity that puts the cell cycle on hold. J. Cell Sci. 2006, 119, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Posada, J.; Yew, N.; Ahn, N.G.; Vande Woude, G.F.; Cooper, J.A. Mos stimulates MAP kinase in Xenopus oocytes and activates a MAP kinase kinase in vitro. Mol. Cell. Biol. 1993, 13, 2546–2553. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, N.; Watanabe, N.; Furuta, Y.; Tamemoto, H.; Sagata, N.; Yokoyama, M.; Okazaki, K.; Nagayoshi, M.; Takeda, N.; Ikawa, Y.; et al. Parthenogenetic activation of c-mos mice. Nature 1994, 370, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.; Rulong, S.; Resau, J.; Fukasawa, K.; Matten, W.; Kuriyama, R.; Mansour, S.; Ahn, N.; Vande Woude, G.F. Mos/mitogen-activated protein kinase can induce early meiotic phenotypes in the absence of maturation-promoting factor: A novel system for analyzing spindle formation during meiosis I. Proc. Natl. Acad. Sci. USA. 1996, 93, 4730–4735. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.D.; Lewellyn, A.L.; Maller, J.L. A Constitutively Active Form of the Protein Kinase p90Rsk1 Is Sufficient to Trigger the G2/M Transition in Xenopus Oocytes. J. Biol. Chem. 2001, 276, 46099–46103. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, R.R.; Ferrell, J.E., Jr. The Protein Kinase p90 Rsk as an Essential Mediator of Cytostatic Factor Activity. Science 2016, 286, 1362–1365. [Google Scholar] [CrossRef]
- Zachariae, W.; Nasmyth, K. Whose end is destruction: Cell division and the anaphase- promoting complex. Genes Dev. 1999, 13, 2039–2058. [Google Scholar] [CrossRef] [PubMed]
- Prinz, S.; Hwang, E.S.; Visintin, R.; Amon, A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 1998, 8, 750–760. [Google Scholar] [CrossRef]
- Fang, G.; Yu, H.; Kirschner, M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998, 12, 1871–1883. [Google Scholar] [CrossRef] [PubMed]
- Vanoosthuyse, V.; Valsdottir, R.; Javerzat, J.-P.; Hardwick, K.G. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol. Cell. Biol. 2004, 24, 9786–9801. [Google Scholar] [CrossRef] [PubMed]
- Maller, J.L.; Schwab, M.S.; Gross, S.D.; Taieb, F.E.; Roberts, B.T.; Tunquist, B.J. The mechanism of CSF arrest in vertebrate oocytes. Mol. Cell. Endocrinol. 2002, 187, 173–178. [Google Scholar] [CrossRef]
- Schwab, M.S.; Roberts, B.T.; Gross, S.D.; Tunquist, B.J.; Taieb, F.E.; Lewellyn, A.L.; Maller, J.L. Bub1 is activated by the protein kinase p90(Rsk) during Xenopus oocyte maturation. Curr. Biol. 2001, 11, 141–150. [Google Scholar] [CrossRef]
- Ni, H.; Sheng, X.; Cui, X.; Gu, M.; Liu, Y.; Qi, X.; Xing, S.; Guo, Y. Epidermal growth factor-mediated mitogen-activated protein Kinase3/1 pathway is conducive to in vitro maturation of sheep oocytes. PLoS ONE 2015, 10, e0120418. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, S.; Labudda, A.; Meinecke, B. In vitro ageing of porcine oocytes: Changes in phosphorylation of the mitogen-activated protein kinase (MAPK) and parthenogenetic activability. Reprod. Domest. Anim. 2010, 45, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, D.; Hou, Y.; Li, Y.-H.; Sun, Q.-Y.; Sun, X.-F.; Wang, W.-H. Reduced expression of MAD2, BCL2, and MAP kinase activity in pig oocytes after in vitro aging are associated with defects in sister chromatid segregation during meiosis II and embryo fragmentation after activation. Biol. Reprod. 2005, 72, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C.; Xiong, B.; Lu, S.S.; Sun, Q.Y. MEK1/2 is a critical regulator of microtubule assembly and spindle organization during rat oocyte meiotic maturation. Mol. Reprod. Dev. 2008, 75, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Miyagaki, Y.; Kanemori, Y.; Baba, T. Possible involvement of mitogen- and stress-activated protein kinase 1, MSK1, in metaphase-II arrest through phosphorylation of EMI2 in mouse oocytes. Dev. Biol. 2011, 359, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Gupta, A.; Sharma, A.; Prasad, S.; Pandey, A.N.; Yadav, P.K.; Pandey, A.K.; Shrivastav, T.G.; Chaube, S.K. Role of Mitogen Activated Protein Kinase and Maturation Promoting Factor During the Achievement of Meiotic Competency in Mammalian Oocytes. J. Cell. Biochem. 2018, 119, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, E.; Yoshida, N.; Kubo, A.; Li, H.; Okuda, E.; Amanai, M.; Perry, A.C.F. Mouse Emi2 as a distinctive regulatory hub in second meiotic metaphase. Development 2010, 137, 3281–3291. [Google Scholar] [CrossRef] [PubMed]
- Munroe, D.; Jacobson, A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 1990, 10, 3441–3455. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005, 433, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Hake, L.E.; Andresson, T.; Littlepage, L.E.; Ruderman, J.V.; Richter, J.D. Phosphorylation of CPE binding factor by Eg2 regulates translation of c- mos mRNA. Nature 2000, 404, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.; Raught, B.; Sonenberg, N. eIF4 Initiation Factors: Effectors of mRNA Recruitment to Ribosomes and Regulators. Annu. Rev. Biochem. 1999, 68, 913–963. [Google Scholar] [CrossRef] [PubMed]
- Pyronnet, S.; Imataka, H.; Gingras, A.C.; Fukunaga, R.; Hunter, T.; Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999, 18, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Harms, U.; Andreou, A.Z.; Gubaev, A.; Klostermeier, D. EIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 2014, 42, 7911–7922. [Google Scholar] [CrossRef] [PubMed]
- Etchison, D.; Milburn, S.C.; Edery, I.; Sonenberg, N.; Hershey, J.W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotc initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982, 257, 14806–14810. [Google Scholar] [PubMed]
- Pain, V.M. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996, 236, 747–771. [Google Scholar] [CrossRef] [PubMed]
- Méthot, N.; Song, M.S.; Sonenberg, N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 1996, 16, 5328–5334. [Google Scholar] [CrossRef] [PubMed]
- Vornlocher, H.; Hanachi, P.; Ribeiro, S. A 110-Kilodalton Subunit of Translation Initiation Factor eIF3 and an Associated 135-kilodalton Protein Are Encoded by theSaccharomyces cerevisiae TIF32 and TIF31Genes. J. Biol. Chem. 1999, 274, 16802–16812. [Google Scholar] [CrossRef] [PubMed]
- Pause, A.; Belsham, G.J.; Gingras, A.-C.; Donzé, O.; Lin, T.-A.; Lawrence, J.C.; Sonenberg, N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 1994, 371, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Josse, L.; Xie, J.; Proud, C.G.; Smales, C.M. mTORC1 signalling and eIF4E/4E-BP1 translation initiation factor stoichiometry influence recombinant protein productivity from GS-CHOK1 cells. Biochem. J. 2016, 473, 4651–4664. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Raught, B.; Gygi, S.P.; Niedzwiecka, A.; Miron, M.; Burley, S.K.; Polakiewicz, R.D.; Wyslouch-Cieszynska, A.; Aebersold, R.; Sonenberg, N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001, 15, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Vander Haar, E.; Lee, S.I.; Bandhakavi, S.; Griffin, T.J.; Kim, D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007, 9, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cocrystal Structure of the Messenger RNA 5′ Cap-Binding Protein (eIF4E) Bound to 7-methyl-GDP. Cell 1997, 89, 951–961. [Google Scholar] [CrossRef]
- Matsuo, H.; Li, H.; McGuire, A.M.; Mark Fletcher, C.; Gingras, A.C.; Sonenberg, N.; Wagner, G. Structure of translation factor elF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 1997, 4, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Scheper, G.C.; Proud, C.G. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 2002, 269, 5350–5359. [Google Scholar] [CrossRef] [PubMed]
- Slepenkov, S.V.; Darzynkiewicz, E.; Rhoads, R.E. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: Evidence for a one-step binding mechanism. J. Biol. Chem. 2006, 281, 14927–14938. [Google Scholar] [CrossRef] [PubMed]
- Morley, S.J.; Naegele, S. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J. Biol. Chem. 2002, 277, 32855–32859. [Google Scholar] [CrossRef] [PubMed]
- Zuberek, J.; Jemielity, J.; Jablonowska, A.; Stepinski, J.; Dadlez, M.; Stolarski, R.; Darzynkiewicz, E. Influence of Electric Charge Variation at Residues 209 and 159 on the Interaction of eIF4E with the mRNA 5′ Terminus. Biochemistry 2004, 43, 5370–5379. [Google Scholar] [CrossRef] [PubMed]
- Dowling, R.; Topisirovic, I.; Alain, T.; Bidinosti, M. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs Supplemental data. Science 2010, 328, 1172–1176. [Google Scholar] [CrossRef] [PubMed]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, D.; Mohr, I. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev. 2014, 28, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Naegele, S.; Morley, S.J. Molecular cross-talk between MEK1/2 and mTOR signalling during recovery of 293 cells from hypertonic stress. J. Biol. Chem. 2004, 279, 46023–46034. [Google Scholar] [CrossRef] [PubMed]
- McGrew, L.L.; Dworkin-Rastl, E.; Dworkin, M.B.; Richter, J.D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989, 3, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.A.; Sheets, M.D.; Wickens, M.P. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989, 3, 2151–2162. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Swenson, K.; Piwnica-Worms, H.; Richter, J.D. Maturation-specific polyadenylation: In vitro activation by p34(cdc2) and phosphorylation of a 58-kD CPE-binding protein. Genes Dev. 1991, 5, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Hake, L.E.; Richter, J.D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 1994, 79, 617–627. [Google Scholar] [CrossRef]
- Mendez, R.; Barnard, D.; Richter, J.D. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002, 21, 1833–1844. [Google Scholar] [CrossRef] [PubMed]
- Hodgman, R.; Tay, J.; Mendez, R.; Richter, J.D. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development 2001, 128, 2815–2822. [Google Scholar] [PubMed]
- Tomek, W.; Wollenhaupt, K. The “closed loop model” in controlling mRNA translation during development. Anim. Reprod. Sci. 2012, 134, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Siemer, C.; Smiljakovic, T.; Bhojwani, M.; Leiding, C.; Kanitz, W.; Kubelka, M.; Tomek, W. Analysis of mRNA associated factors during bovine oocyte maturation and early embryonic development. Mol. Reprod. Dev. 2009, 76, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Ellederova, Z.; Kovarova, H.; Melo-Sterza, F.; Livingstone, M.; Tomek, W.; Kubelka, M. Suppression of translation during in vitro maturation of pig oocytes despite enhanced formation of cap-binding protein complex eIF4F and 4E-BP1 hyperphosphorylation. Mol. Reprod. Dev. 2006, 73, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Tomek, W.; Melo Sterza, F.A.; Kubelka, M.; Wollenhaupt, K.; Torner, H.; Anger, M.; Kanitz, W. Regulation of translation during in vitro maturation of bovine oocytes: The role of MAP kinase, eIF4E (cap binding protein) phosphorylation, and eIF4E-BP1. Biol. Reprod. 2002, 66, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Susor, A.; Jansova, D.; Cerna, R.; Danylevska, A.; Anger, M.; Toralova, T.; Malik, R.; Supolikova, J.; Cook, M.S.; Oh, J.S.; et al. Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jansova, D.; Koncicka, M.; Tetkova, A.; Cerna, R.; Malik, R.; del Llano, E.; Kubelka, M.; Susor, A. Regulation of 4E-BP1 activity in the mammalian oocyte. Cell Cycle 2017, 16, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Šušor, A.; Jelínková, L.; Karabínová, P.; Torner, H.; Tomek, W.; Kovářová, H.; Kubelka, M. Regulation of cap-dependent translation initiation in the early stage porcine parthenotes. Mol. Reprod. Dev. 2008, 75, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.; Wrenzycki, C.; Tomek, W. Inactivation of mTor arrests bovine oocytes in the metaphase-I stage, despite reversible inhibition of 4E-BP1 phosphorylation. Mol. Reprod. Dev. 2014, 81, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Severance, A.L.; Latham, K.E. PLK1 regulates spindle association of phosphorylated eukaryotic translation initiation factor 4E binding protein, and spindle function in mouse oocytes. Am. J. Physiol. Cell Physiol. 2017, 313, C501–C515. [Google Scholar] [CrossRef] [PubMed]
- Lapasset, L.; Pradet-Balade, B.; Vergé, V.; Lozano, J.C.; Oulhen, N.; Cormier, P.; Peaucellier, G. Cyclin B synthesis and rapamycin-sensitive regulation of protein synthesis during starfish oocyte meiotic divisions. Mol. Reprod. Dev. 2008, 75, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Hunter, T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997, 16, 1921–1933. [Google Scholar] [CrossRef] [PubMed]
- Messina, V.; Di Sauro, A.; Pedrotti, S.; Adesso, L.; Latina, A.; Geremia, R.; Rossi, P.; Sette, C. Differential contribution of the MTOR and MNK pathways to the regulation of mRNA translation in meiotic and postmeiotic mouse male germ cells. Biol. Reprod. 2010, 83, 607–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yue, P.; Deng, X.; Ueda, T.; Fukunaga, R.; Khuri, F.R.; Sun, S.-Y. Protein Phosphatase 2A Negatively Regulates Eukaryotic Initiation Factor 4E Phosphorylation and eIF4F Assembly through Direct Dephosphorylation of Mnk and eIF4E. Neoplasia 2010, 12, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Shveygert, M.; Kaiser, C.; Bradrick, S.S.; Gromeier, M. Regulation of Eukaryotic Initiation Factor 4E (eIF4E) Phosphorylation by Mitogen-Activated Protein Kinase Occurs through Modulation of Mnk1-eIF4G Interaction. Mol. Cell. Biol. 2010, 30, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Ellederová, Z.; Cais, O.; Šušor, A.; Uhlířová, K.; Kovářová, H.; Jelínková, L.; Tomek, W.; Kubelka, M. ERK1/2 map kinase metabolic pathway is responsible for phosphorylation of translation initiation factor eIF4E during in vitro maturation of pig oocytes. Mol. Reprod. Dev. 2008, 75, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Brook, M.; Gray, N.K. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem. Soc. Trans. 2012, 40, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic Polyadenylation Element Binding Proteins in Development, Health, and Disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415. [Google Scholar] [CrossRef] [PubMed]
- Huarte, J.; Stutz, A.; O’Connell, M.L.; Gubler, P.; Belin, D.; Darrow, A.L.; Strickland, S.; Vassalli, J.D. Transient translational silencing by reversible mRNA deadenylation. Cell 1992, 69, 1021–1030. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kano, K.; Naito, K. Porcine CPEB1 is involved in Cyclin B translation and meiotic resumption in porcine oocytes. Anim. Sci. J. 2010, 81, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Richter, J.D. Opposing Polymerase-Deadenylase Activities Regulate Cytoplasmic Polyadenylation. Mol. Cell 2006, 24, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Miranda, G.; Méndez, R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res. Rev. 2012, 11, 460–472. [Google Scholar] [CrossRef] [PubMed]
- de Moor, C.H.; Richter, J.D. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 1997, 17, 6419–6426. [Google Scholar] [CrossRef] [PubMed]
- Brunet, S.; Dumont, J.; Lee, K.W.; Kinoshita, K.; Hikal, P.; Gruss, O.J.; Maro, B.; Verlhac, M.H. Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS ONE 2008, 3, e3338. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ji, S.Y.; Sha, Q.Q.; Dang, Y.; Zhou, J.J.; Zhang, Y.L.; Liu, Y.; Wang, Z.W.; Hu, B.; Sun, Q.Y.; et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat. Struct. Mol. Biol. 2016, 23, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Setoyama, D.; Yamashita, M.; Sagata, N. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 18001–18006. [Google Scholar] [CrossRef] [PubMed]
- Keady, B.T.; Kuo, P.; Martínez, S.E.; Yuan, L.; Hake, L.E. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J. Cell Sci. 2007, 120, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Komrskova, P.; Susor, A.; Malik, R.; Prochazkova, B.; Liskova, L.; Supolikova, J.; Hladky, S.; Kubelka, M. Aurora kinase A is not involved in CPEB1 phosphorylation and cyclin B1 mRNA polyadenylation during meiotic maturation of porcine oocytes. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Sha, Q.-Q.; Dai, X.-X.; Dang, Y.; Tang, F.; Liu, J.; Zhang, Y.-L.; Fan, H.-Y. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 2017, 144, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Mulner-Lorillon, O.; Chassé, H.; Morales, J.; Bellé, R.; Cormier, P. MAPK/ERK activity is required for the successful progression of mitosis in sea urchin embryos. Dev. Biol. 2017, 421, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Kracmarova, J. Role of MAPK in Regulation of Cytoplasmic Polyadenylation during Meiotic Maturation of Mammalian Oocytes; University Charles: Prague, Czech Republic, 2017. [Google Scholar]
- Martins, J.P.S.; Liu, X.; Oke, A.; Arora, R.; Franciosi, F.; Viville, S.; Laird, D.J.; Fung, J.C.; Conti, M. DAZL and CPEB1 regulate mRNA translation synergistically during oocyte maturation. J. Cell Sci. 2016, 129, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, C.G.; Maines, J.Z.; Wasserman, S.A. Meiotic cell cycle requirement for a fly homologue of human deleted in Azoospermia. Nature 1996, 381, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Ruggiu, M.; Speed, R.; Taggart, M.; McKay, S.J.; Kilanowski, F.; Saunders, P.; Dorin, J.; Cooke, H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997, 389, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Eckerdt, F.; Pascreau, G.; Phistry, M.; Lewellyn, A.L.; DePaoli-Roach, A.A.; Maller, J.L. Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle 2009, 8, 2413–2419. [Google Scholar] [CrossRef] [PubMed]
- Helmke, K.J.; Heald, R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 2014, 206, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, A.; Loiarro, M.; Bielli, P.; Busà, R.; Paronetto, M.P.; Loreni, F.; Geremia, R.; Sette, C. Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis 2008, 29, 2279–2288. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases A large group of genes in all eukaryotes encode for. October 2000, 103, 211–225. [Google Scholar] [CrossRef]
- Karar, J.; Maity, A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front. Mol. Neurosci. 2011, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. TRENDS Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Lehr, S.; Kotzka, J.; Avci, H.; Sickmann, A.; Meyer, H.E.; Herkner, A.; Muller-Wieland, D. Identification of major ERK-related phosphorylation sites in Gab1. Biochemistry 2004, 43, 12133–12140. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.J.; Waterfield, M.D.; Downward, J. Phosphatidylinositol-3-OH kinase direct target of Ras. Nature 1994, 370, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, A.S.; Meikle, S.; Yazici, Z.; Eulitz, M.; Kolch, W. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 2002, 21, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Nakdimon, I.; Walser, M.; Fröhli, E.; Hajnal, A. PTEN Negatively Regulates MAPK Signaling during Caenorhabditis elegans Vulval Development. PLoS Genet. 2012, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKay, M.M.; Morrison, D.K. Integrating signals from RTKs to ERK/MAPK. Oncogene 2007, 26, 3113–3121. [Google Scholar] [CrossRef] [PubMed]
- Fissore, R.A.; He, C.L.; Vande Woude, G.F. Potential role of mitogen-activated protein kinase during meiosis resumption in bovine oocytes. Biol. Reprod. 1996, 55, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Duncan, F.E.; Jasti, S.; Paulson, A.; Kelsh, J.M.; Fegley, B.; Gerton, J.L. Age-associated dysregulation of protein metabolism in the mammalian oocyte. Aging Cell 2017, 16, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.A.; Yoon, Y.J.; Singer, R.H. Imaging translation in single cells using fluorescent microscopy. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Romasko, E.J.; Amarnath, D.; Midic, U.; Latham, K.E. Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: Localized translational control supporting female meiosis in mammals. Genetics 2013, 195, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Sabatini, D.M. Defining the Role of mTOR in Cancer. Cancer Cell 2007, 12, 9–22. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalous, J.; Tetkova, A.; Kubelka, M.; Susor, A. Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis. Int. J. Mol. Sci. 2018, 19, 698. https://doi.org/10.3390/ijms19030698
Kalous J, Tetkova A, Kubelka M, Susor A. Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis. International Journal of Molecular Sciences. 2018; 19(3):698. https://doi.org/10.3390/ijms19030698
Chicago/Turabian StyleKalous, Jaroslav, Anna Tetkova, Michal Kubelka, and Andrej Susor. 2018. "Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis" International Journal of Molecular Sciences 19, no. 3: 698. https://doi.org/10.3390/ijms19030698
APA StyleKalous, J., Tetkova, A., Kubelka, M., & Susor, A. (2018). Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis. International Journal of Molecular Sciences, 19(3), 698. https://doi.org/10.3390/ijms19030698