Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions
Abstract
:1. Introduction
2. Results
2.1. Decreased Numbers of Cortical Iba1+ Microglia in Aged Mice
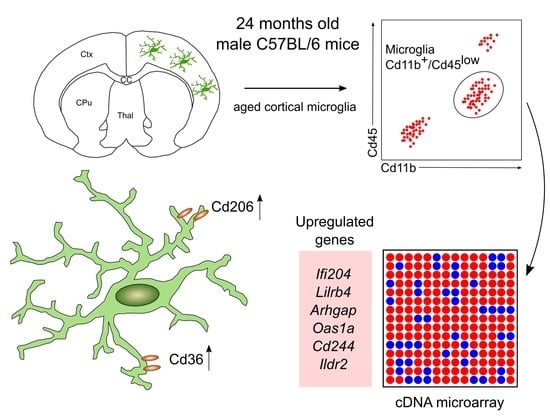
2.2. Detection of a Distinct Gene Expression Profile in Aged Cortical Microglia
2.3. Increased Expression of Cd206 and Cd36 in Aged Cortical Microglia
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Microglia Isolation
4.3. Flow Cytometry
4.4. Histology and Immunohistochemistry
4.5. Determination of Cortical Thickness and Cortical Microglia Numbers
4.6. cDNA Microarray
4.7. Statistics
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McHugh, D.; Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 2018, 217, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Molteni, M.; Rossetti, C. Neurodegenerative diseases: The immunological perspective. J. Neuroimmunol. 2017, 313, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C.; et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013, 16, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Greter, M.; Lelios, I.; Pelczar, P.; Hoeffel, G.; Price, J.; Leboeuf, M.; Kündig, T.M.; Frei, K.; Ginhoux, F.; Merad, M.; et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity 2012, 37, 1050–1060. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Askew, K.; Li, K.; Olmos-Alonso, A.; Garcia-Moreno, F.; Liang, Y.; Richardson, P.; Tipton, T.; Chapman, M.A.; Riecken, K.; Beccari, S.; et al. Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Rep. 2017, 18, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Ajami, B.; Bennett, J.L.; Krieger, C.; Tetzlaff, W.; Rossi, F.M.V. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007, 10, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, T.; Wieghofer, P.; Jordão, M.J.C.; Prutek, F.; Hagemeyer, N.; Frenzel, K.; Amann, L.; Staszewski, O.; Kierdorf, K.; Krueger, M.; et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 2016, 17, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Spittau, B. Aging Microglia-Phenotypes, Functions and Implications for Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2017, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Raj, D.D.A.; Jaarsma, D.; Holtman, I.R.; Olah, M.; Ferreira, F.M.; Schaafsma, W.; Brouwer, N.; Meijer, M.M.; de Waard, M.C.; van der Pluijm, I.; et al. Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol. Aging 2014. [Google Scholar] [CrossRef] [PubMed]
- Raj, D.D.A.; Moser, J.; van der Pol, S.M.A.; van Os, R.P.; Holtman, I.R.; Brouwer, N.; Oeseburg, H.; Schaafsma, W.; Wesseling, E.M.; den Dunnen, W.; et al. Enhanced microglial pro-inflammatory response to lipopolysaccharide correlates with brain infiltration and blood-brain barrier dysregulation in a mouse model of telomere shortening. Aging Cell 2015, 14, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Gottfried-Blackmore, A.C.; McEwen, B.S.; Bulloch, K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia 2007, 55, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Facci, L.; Barbierato, M.; Marinelli, C.; Argentini, C.; Skaper, S.D.; Giusti, P. Toll-like receptors 2, -3 and -4 prime microglia but not astrocytes across central nervous system regions for ATP-dependent interleukin-1β release. Sci. Rep. 2014, 4, 6824. [Google Scholar] [CrossRef] [PubMed]
- Fonken, L.K.; Frank, M.G.; Kitt, M.M.; D’Angelo, H.M.; Norden, D.M.; Weber, M.D.; Barrientos, R.M.; Godbout, J.P.; Watkins, L.R.; Maier, S.F. The Alarmin HMGB1 Mediates Age-Induced Neuroinflammatory Priming. J. Neurosci. 2016, 36, 7946–7956. [Google Scholar] [CrossRef] [PubMed]
- Holtman, I.R.; Raj, D.D.; Miller, J.A.; Schaafsma, W.; Yin, Z.; Brouwer, N.; Wes, P.D.; Möller, T.; Orre, M.; Kamphuis, W.; et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 2015, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.-C.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharaf, A.; Krieglstein, K.; Spittau, B. Distribution of microglia in the postnatal murine nigrostriatal system. Cell Tissue Res. 2013, 351, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.S.; Ma, J.; Jegathees, T.; Goldsbury, C. Microglia show altered morphology and reduced arborization in human brain during aging and Alzheimer’s disease. Brain Pathol. 2017, 27, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Koellhoffer, E.C.; McCullough, L.D.; Ritzel, R.M. Old Maids: Aging and Its Impact on Microglia Function. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Dauffy, J.; Mouchiroud, G.; Bourette, R.P. The interferon-inducible gene, Ifi204, is transcriptionally activated in response to M-CSF, and its expression favors macrophage differentiation in myeloid progenitor cells. J. Leukoc. Biol. 2006, 79, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Chunfa, L.; Xin, S.; Qiang, L.; Sreevatsan, S.; Yang, L.; Zhao, D.; Zhou, X. The Central Role of IFI204 in IFN-β Release and Autophagy Activation during Mycobacterium bovis Infection. Front. Cell. Infect. Microbiol. 2017, 7, 169. [Google Scholar] [CrossRef] [PubMed]
- Blank, T.; Prinz, M. Type I interferon pathway in CNS homeostasis and neurological disorders. Glia 2017, 65, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Mohammed, F.; Nam, G.; Chen, Y.; Qi, J.; Garner, L.I.; Allen, R.L.; Yan, J.; Willcox, B.E.; Gao, G.F. Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): A myeloid inhibitory receptor involved in immune tolerance. J. Biol. Chem. 2011, 286, 18013–18025. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Qin, J.-J.; Zhang, Y.; Cheng, W.-L.; Ji, Y.-X.; Gong, F.-H.; Zhu, X.-Y.; Zhang, Y.; She, Z.-G.; Huang, Z.; et al. LILRB4 deficiency aggravates the development of atherosclerosis and plaque instability by increasing the macrophage inflammatory response via NF-κB signaling. Clin. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.S.; Friend, D.S.; Lee, D.M.; Li, L.; Austen, K.F.; Katz, H.R. gp49B1 deficiency is associated with increases in cytokine and chemokine production and severity of proliferative synovitis induced by anti-type II collagen mAb. Eur. J. Immunol. 2005, 35, 1530–1538. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, W.; Kooijman, L.; Schetters, S.; Orre, M.; Hol, E.M. Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta 2016, 1862, 1847–1860. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, V.; Armentano, M.; Sarò, G.; Ciraolo, E.; Ghigo, A.; Germena, G.; Umbach, A.; Valnegri, P.; Passafaro, M.; Carabelli, V.; et al. Disruption of ArhGAP15 results in hyperactive Rac1, affects the architecture and function of hippocampal inhibitory neurons and causes cognitive deficits. Sci. Rep. 2016, 6, 34877. [Google Scholar] [CrossRef] [PubMed]
- Bedard, K.; Krause, K.-H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Spencer, N.Y.; Pantazis, N.J.; Engelhardt, J.F. Alsin and SOD1(G93A) proteins regulate endosomal reactive oxygen species production by glial cells and proinflammatory pathways responsible for neurotoxicity. J. Biol. Chem. 2011, 286, 40151–40162. [Google Scholar] [CrossRef] [PubMed]
- Georgoudaki, A.-M.; Khodabandeh, S.; Puiac, S.; Persson, C.M.; Larsson, M.K.; Lind, M.; Hammarfjord, O.; Nabatti, T.H.; Wallin, R.P.A.; Yrlid, U.; et al. CD244 is expressed on dendritic cells and regulates their functions. Immunol. Cell Biol. 2015, 93, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Porrini, V.; Lanzillotta, A.; Branca, C.; Benarese, M.; Parrella, E.; Lorenzini, L.; Calzà, L.; Flaibani, R.; Spano, P.F.; Imbimbo, B.P.; et al. CHF5074 (CSP-1103) induces microglia alternative activation in plaque-free Tg2576 mice and primary glial cultures exposed to beta-amyloid. Neuroscience 2015, 302, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Yu, W.; Tian, Q.; Fu, X.; Wang, X.; Gu, M.; Lü, Y. Chitinase1 contributed to a potential protection via microglia polarization and Aβ oligomer reduction in d-galactose and aluminum-induced rat model with cognitive impairments. Neuroscience 2017, 355, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-M.; Mun, B.-R.; Lee, S.-J.; Joh, Y.; Lee, H.-Y.; Ji, K.-Y.; Choi, H.-R.; Lee, E.-H.; Kim, E.-M.; Jang, J.-H.; et al. TREM2 promotes Aβ phagocytosis by upregulating C/EBPα-dependent CD36 expression in microglia. Sci. Rep. 2017, 7, 11118. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Faustino, J.; Woo, M.-S.; Derugin, N.; Vexler, Z.S. Lack of the scavenger receptor CD36 alters microglial phenotypes after neonatal stroke. J. Neurochem. 2015, 135, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Woo, M.-S.; Wang, X.; Faustino, J.V.; Derugin, N.; Wendland, M.F.; Zhou, P.; Iadecola, C.; Vexler, Z.S. Genetic deletion of CD36 enhances injury after acute neonatal stroke. Ann. Neurol. 2012, 72, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Ueno, M.; Fujita, Y.; Tanaka, T.; Nakamura, Y.; Kikuta, J.; Ishii, M.; Yamashita, T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013, 16, 543–551. [Google Scholar] [CrossRef] [PubMed]
- De Haas, A.H.; Boddeke, H.W.G.M.; Biber, K. Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia 2008, 56, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Olah, M.; Patrick, E.; Villani, A.-C.; Xu, J.; White, C.C.; Ryan, K.J.; Piehowski, P.; Kapasi, A.; Nejad, P.; Cimpean, M.; et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 2018, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Mrdjen, D.; Pavlovic, A.; Hartmann, F.J.; Schreiner, B.; Utz, S.G.; Leung, B.P.; Lelios, I.; Heppner, F.L.; Kipnis, J.; Merkler, D.; et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 2018. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.J.; Dimmic, M.W. A two-sample Bayesian t-test for microarray data. BMC Bioinform. 2006, 7, 126. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zöller, T.; Attaai, A.; Potru, P.S.; Ruß, T.; Spittau, B. Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions. Int. J. Mol. Sci. 2018, 19, 706. https://doi.org/10.3390/ijms19030706
Zöller T, Attaai A, Potru PS, Ruß T, Spittau B. Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions. International Journal of Molecular Sciences. 2018; 19(3):706. https://doi.org/10.3390/ijms19030706
Chicago/Turabian StyleZöller, Tanja, Abdelraheim Attaai, Phani Sankar Potru, Tamara Ruß, and Björn Spittau. 2018. "Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions" International Journal of Molecular Sciences 19, no. 3: 706. https://doi.org/10.3390/ijms19030706
APA StyleZöller, T., Attaai, A., Potru, P. S., Ruß, T., & Spittau, B. (2018). Aged Mouse Cortical Microglia Display an Activation Profile Suggesting Immunotolerogenic Functions. International Journal of Molecular Sciences, 19(3), 706. https://doi.org/10.3390/ijms19030706