Characterization of Multiple Cytokine Combinations and TGF-β on Differentiation and Functions of Myeloid-Derived Suppressor Cells
Abstract
:1. Introduction
2. Results
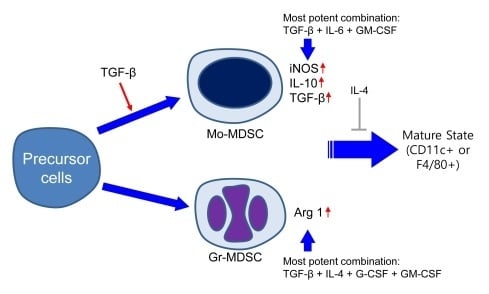
2.1. Characterization of Subpopulation in the Differentiated MDSCs Induced by Different Cytokine Combinations with or without TGF-β
2.2. Characterization of Immature State of Differentiated MDSCs Induced by Different Cytokine Combinations with or without TGF-β
2.3. MDSCs Derived Using Different Cytokine Combinations Express Immunosuppressive Molecules, the Expression of Which is Enhanced by TGF-β
2.4. MDSCs Derived Using Different Cytokine Combinations Suppress T Cell Proliferation, an Activity Enhanced by TGF-β
3. Discussion
4. Materials and Methods
4.1. Mice and MDSC Isolation
4.2. Flow Cytometry
4.3. Quantitative Reverse Transcription-Polymerase Chain Reaction
4.4. In Vitro Differentiation of Bone Marrow-Derived MDSCs
4.5. In Vitro Suppression Assays
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MDSCs | myeloid-derived suppressor cells |
| Gr-MDSC | granulocytic-MDSC |
| Mo-MDSC | monocytic-MDSC |
| ROS | reactive oxygen species |
| Arg1 | arginase 1 |
| NO | nitric oxide |
| iNOS | inducible nitric oxide synthase |
| TGF-β | transforming growth factor-β |
| IL-10 | interleukin-10 |
| CSF | colony-stimulating factors |
| GM-CSF | granulocyte-macrophage CSF |
| M-CSF | macrophage CSF |
| G-CSF | granulocyte CSF |
| STAT3 | signal transducer and activator of transcription 3 |
| IL-4Rα | interleukin-4 receptor α |
| IL-4 | interleukin-4 |
| IL-6 | interleukin-6 |
| LPS | lipopolysaccharide |
| PE | phycoerythrin |
| APC | allophycocyanin |
| PCR | polymerase chain reaction |
References
- Serafini, P.; Borrello, I.; Bronte, V. Myeloid suppressor cells in cancer: Recruitment, phenotype, properties, and mechanisms of immune suppression. Semin. Cancer Biol. 2006, 16, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Marigo, I.; Dolcetti, L.; Serafini, P.; Zanovello, P.; Bronte, V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol. Rev. 2008, 222, 162–179. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V. Myeloid-derived suppressor cells in inflammation: Uncovering cell subsets with enhanced immunosuppressive functions. Eur. J. Immunol. 2009, 39, 2670–2672. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.; Kim, H.E.; Park, S.G. Insights into Myeloid-Derived Suppressor Cells in Inflammatory Diseases. Arch. Immunol. Ther. Exp. 2015, 63, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.I.; Nagaraj, S.; Collazo, M.; Gabrilovich, D.I. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008, 181, 5791–5802. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Pan, P.Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.H. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.A.; Achuthan, A. Colony stimulating factors and myeloid cell biology in health and disease. Trends Immunol. 2013, 34, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Parmiani, G.; Castelli, C.; Pilla, L.; Santinami, M.; Colombo, M.P.; Rivoltini, L. Opposite immune functions of GM-CSF administered as vaccine adjuvant in cancer patients. Ann. Oncol. 2007, 18, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Serafini, P.; Carbley, R.; Noonan, K.A.; Tan, G.; Bronte, V.; Borrello, I. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res. 2004, 64, 6337–6343. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.K.; Kmieciak, M.; Knutson, K.L.; Bear, H.D.; Manjili, M.H. GM-CSF is one of the main breast tumor-derived soluble factors involved in the differentiation of CD11b-Gr1- bone marrow progenitor cells into myeloid-derived suppressor cells. Breast Cancer Res. Treat. 2010, 123, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Sawanobori, Y.; Ueha, S.; Kurachi, M.; Shimaoka, T.; Talmadge, J.E.; Abe, J.; Shono, Y.; Kitabatake, M.; Kakimi, K.; Mukaida, N.; et al. Chemokine-mediated rapid turnover of myeloid-derived suppressor cells in tumor-bearing mice. Blood 2008, 111, 5457–5466. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Grail, D.; Hodgson, G.; Metcalf, D.; Stanley, E.; Cheers, C.; Fowler, K.J.; Basu, S.; Zhan, Y.F.; Dunn, A.R. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 1994, 84, 1737–1746. [Google Scholar] [PubMed]
- Li, W.; Zhang, X.; Chen, Y.; Xie, Y.; Liu, J.; Feng, Q.; Wang, Y.; Yuan, W.; Ma, J. G-CSF is a key modulator of MDSC and could be a potential therapeutic target in colitis-associated colorectal cancers. Protein Cell 2016, 7, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Gallina, G.; Dolcetti, L.; Serafini, P.; De Santo, C.; Marigo, I.; Colombo, M.P.; Basso, G.; Brombacher, F.; Borrello, I.; Zanovello, P.; et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Investig. 2006, 116, 2777–2790. [Google Scholar] [CrossRef] [PubMed]
- Schnyder, B.; Lugli, S.; Feng, N.; Etter, H.; Lutz, R.A.; Ryffel, B.; Sugamura, K.; Wunderli-Allenspach, H.; Moser, R. Interleukin-4 (IL-4) and IL-13 bind to a shared heterodimeric complex on endothelial cells mediating vascular cell adhesion molecule-1 induction in the absence of the common gamma chain. Blood 1996, 87, 4286–4295. [Google Scholar] [PubMed]
- Mandruzzato, S.; Solito, S.; Falisi, E.; Francescato, S.; Chiarion-Sileni, V.; Mocellin, S.; Zanon, A.; Rossi, C.R.; Nitti, D.; Bronte, V.; et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J. Immunol. 2009, 182, 6562–6568. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Serafini, P.; De Santo, C.; Marigo, I.; Tosello, V.; Mazzoni, A.; Segal, D.M.; Staib, C.; Lowel, M.; Sutter, G.; et al. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J. Immunol. 2003, 170, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Apolloni, E.; Bronte, V.; Mazzoni, A.; Serafini, P.; Cabrelle, A.; Segal, D.M.; Young, H.A.; Zanovello, P. Immortalized myeloid suppressor cells trigger apoptosis in antigen-activated T lymphocytes. J. Immunol. 2000, 165, 6723–6730. [Google Scholar] [CrossRef] [PubMed]
- Bunt, S.K.; Yang, L.; Sinha, P.; Clements, V.K.; Leips, J.; Ostrand-Rosenberg, S. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. 2007, 67, 10019–10026. [Google Scholar] [CrossRef] [PubMed]
- Tamadaho, R.S.E.; Hoerauf, A.; Layland, L.E. Immunomodulatory effects of myeloid-derived suppressor cells in diseases: role in cancer and infections. Immunobiology 2017, 222, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Kwak, Y.; Yang, T.; Han, J.H.; Park, S.H.; Ye, M.B.; Lee, W.; Sim, K.Y.; Kang, J.A.; Kim, Y.C.; et al. Myeloid-Derived Suppressor Cells Are Controlled by Regulatory T Cells via TGF-beta during Murine Colitis. Cell Rep. 2016, 17, 3219–3232. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.G.; Liebertz, D.J.; Epstein, A.L. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J. Immunol. 2010, 185, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Hodgson, G.; Katz, M.; Dunn, A.R. Evaluation of role of G-CSF in the production, survival, and release of neutrophils from bone marrow into circulation. Blood 2002, 100, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Drujont, L.; Carretero-Iglesia, L.; Bouchet-Delbos, L.; Beriou, G.; Merieau, E.; Hill, M.; Delneste, Y.; Cuturi, M.C.; Louvet, C. Evaluation of the therapeutic potential of bone marrow-derived myeloid suppressor cell (MDSC) adoptive transfer in mouse models of autoimmunity and allograft rejection. PLoS ONE 2014, 9, e100013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Highfill, S.L.; Rodriguez, P.C.; Zhou, Q.; Goetz, C.A.; Koehn, B.H.; Veenstra, R.; Taylor, P.A.; Panoskaltsis-Mortari, A.; Serody, J.S.; et al. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood 2010, 116, 5738–5747. [Google Scholar] [CrossRef] [PubMed]
- Lindau, D.; Gielen, P.; Kroesen, M.; Wesseling, P.; Adema, G.J. The immunosuppressive tumour network: Myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 2013, 138, 105–115. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | Mo-MDSCs | Gr-MDSDCs | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM-CSF/IL-4 | GM-CSF/IL-6 | GM-CSF | GM-CSF/G-CSF | GM-CSF/IL-4/G-CSF | GM-CSF/IL-6/G-CSF | GM-CSF/IL-4 | GM-CSF/IL-6 | GM-CSF | GM-CSF/G-CSF | GM-CSF/IL-4/G-CSF | GM-CSF/IL-6//G-CSF | ||||||||||||||
| TGF-β | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | |
| Population change | Percentage | 1 | 5 | 2 | 7 | 2 | 7 | 6 | 9 | 1 | 3 | 6 | 10 | 8 | 3 | 10 | 5 | 9 | 3 | 5 | 1 | 4 | 1 | 4 | 1 |
| Number | 1 | 3 | 1 | 5 | 2 | 6 | 3 | 10 | 1 | 4 | 3 | 10 | 7 | 2 | 9 | 6 | 9 | 5 | 5 | 3 | 3 | 1 | 10 | 3 | |
| Immature Population | F4/80− | 7 | 10 | 3 | 3 | 4 | 1 | 4 | 4 | 7 | 9 | 4 | 4 | 9 | 10 | 8 | 2 | 9 | 1 | 8 | 1 | 9 | 9 | 7 | 1 |
| CD11c− | 9 | 8 | 1 | 1 | 3 | 1 | 4 | 3 | 10 | 9 | 5 | 4 | 9 | 10 | 4 | 2 | 4 | 1 | 4 | 1 | 8 | 5 | 4 | 1 | |
| Suppression Marker | Arginase | 1 | 3 | 1 | 3 | 1 | 2 | 1 | 4 | 3 | 10 | 1 | 3 | 2 | 5 | 1 | 4 | 1 | 3 | 1 | 7 | 3 | 10 | 1 | 6 |
| iNOS | 1 | 3 | 2 | 10 | 2 | 7 | 1 | 9 | 1 | 5 | 2 | 7 | |||||||||||||
| TGF-β | 1 | 1 | 3 | 10 | 1 | 3 | 4 | 5 | 1 | 2 | 3 | 7 | |||||||||||||
| IL-10 | 1 | 4 | 1 | 10 | 1 | 5 | 2 | 4 | 1 | 5 | 1 | 9 | |||||||||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.-R.; Lee, W.; Cho, S.K.; Park, S.-G. Characterization of Multiple Cytokine Combinations and TGF-β on Differentiation and Functions of Myeloid-Derived Suppressor Cells. Int. J. Mol. Sci. 2018, 19, 869. https://doi.org/10.3390/ijms19030869
Lee C-R, Lee W, Cho SK, Park S-G. Characterization of Multiple Cytokine Combinations and TGF-β on Differentiation and Functions of Myeloid-Derived Suppressor Cells. International Journal of Molecular Sciences. 2018; 19(3):869. https://doi.org/10.3390/ijms19030869
Chicago/Turabian StyleLee, Cho-Rong, Wongeun Lee, Steve K. Cho, and Sung-Gyoo Park. 2018. "Characterization of Multiple Cytokine Combinations and TGF-β on Differentiation and Functions of Myeloid-Derived Suppressor Cells" International Journal of Molecular Sciences 19, no. 3: 869. https://doi.org/10.3390/ijms19030869
APA StyleLee, C. -R., Lee, W., Cho, S. K., & Park, S. -G. (2018). Characterization of Multiple Cytokine Combinations and TGF-β on Differentiation and Functions of Myeloid-Derived Suppressor Cells. International Journal of Molecular Sciences, 19(3), 869. https://doi.org/10.3390/ijms19030869