Cytochrome P450 CYP6EV11 in Chironomus kiiensis Larvae Involved in Phenol Stress
Abstract
:1. Introduction
2. Results
2.1. cDNA Cloning and Characterization
2.2. Polygenetic Analysis
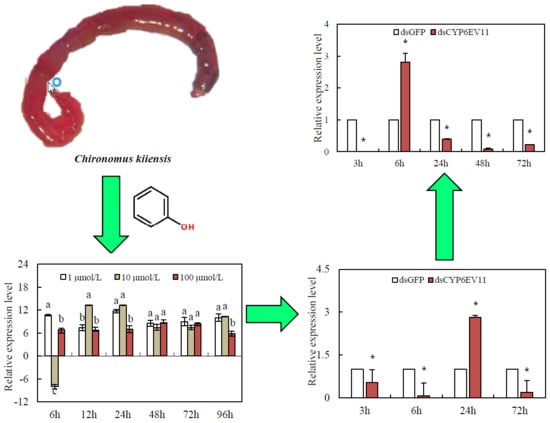
2.3. Expression Profiling under Phenol Stress
2.4. Gene Silencing Analysis
2.5. Effects of Gene Silencing on Development and in Response to Phenol Stress
3. Discussion
4. Materials and Methods
4.1. Experimental Midge Rearing
4.2. Cloning and Identification of CYP6EV11
4.3. Multiple Sequence Alignment and Polygenetic Analysis
4.4. C. kiiensis Larvae Stress and RNAi Analysis
4.5. Real-time RT-PCR Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Antonyraj, C.A.; Srinivasan, K. One-step hydroxylation of benzene to phenol over layered double hydroxides and their derived forms. Catal. Surv. Asia 2013, 17, 47–70. [Google Scholar] [CrossRef]
- Michałowicz, J.; Duda, R.O.W. Analysis of chlorophenols, chlorocatechols, chlorinated methoxyphenols and monoterpenes in communal sewage of ŁÓDŹ and in the Ner River in 1999–2000. Water Air Soil Pollut. 2005, 164, 205–222. [Google Scholar] [CrossRef]
- Dutta, N.N.; Brothakur, S.; Patil, G.S. Phase transfer catalyzed extraction of phenolic substances from aqueous alkaline stream. Sep. Sci. Technol. 1992, 27, 1435–1448. [Google Scholar] [CrossRef]
- Huang, J.H.; Wang, X.G.; Jin, Q.Z.; Liu, Y.F.; Wang, Y. Removal of phenol from aqueous solution by adsorption onto OTMAC-modified attapulgite. J. Environ. Manag. 2007, 84, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Australian Water Research Council (AWRC). Australian Water Quality Criteria for Organic Compounds; Australian Water Research Council Technical Paper. No. 82; AWRC: Canberra, Australia, 1984; Volume 224. [Google Scholar]
- DOE-MU. Water Quality Criteria and Standards for Malaysia. Vol. 4—Criteria and Standards for Organic Constituents; Prepared by Goh, S.H.; Yap, S.Y.; Lim, R.P.; DOE; Malaysia/IPT, University of Malaysia: Kuala Lumpur, Malaysia, 1986; Volume 224. [Google Scholar]
- United States Environmental Protection Agency (USEPA). National Recommended Water Quality Criteria; Office of Water, Office of Science and Technology: Washington, DC, USA, 2009. [Google Scholar]
- Cao, C.W.; Li, X.P.; Ge, S.L.; Sun, L.L.; Wang, Z.Y. Enzymatic activities as potential stress biomarkers of two substituted benzene compounds in Propsilocerus akamusi (Diptera: Chironomidae). Afr. J. Aquat. Sci. 2012, 37, 265–270. [Google Scholar] [CrossRef]
- Nebert, D.W.; Russell, D.W. Clinical importance of the cytochromes P450. Lancet 2002, 360, 1155–1162. [Google Scholar] [CrossRef]
- Liu, X.; Chen, J.; Yang, Z. Characterization and induction of two cytochrome P450 genes, CYP6AE28 and CYP6AE30, in Cnaphalocrocis medinalis: Possible involvement in metabolism of rice allelochemicals. Z. Naturforsch. C 2010, 65, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Poupardin, R.; Riaz, M.A.; Vontas, J.; David, J.P.; Reynaud, S. Transcription profiling of eleven cytochrome P450 s potentially involved in xenobiotic metabolism in the mosquito Aedes aegypti. Insect Mol. Biol. 2010, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, D.; Chynoweth, R.; Guillén, J.; De la Rúa, P.; Bielza, P. Novel cytochrome P450 genes, CYP6EB1 and CYP6EC1, are over-expressed in acrinathrin-resistant Frankliniella occidentalis (Thysanoptera: Thripidae). J. Econ. Entomol. 2012, 105, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Musasia, F.K.; Isaac, A.O.; Masiga, D.K.; Omedo, I.A.; Mwakubambanya, R.; Ochieng, R.; Mireji, P.O. Sex-specific induction of CYP6 cytochrome P450 genes in cadmium and lead tolerant Anopheles gambiae. Malar. J. 2013, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Edi, C.V.; Djogbénou, L.; Jenkins, A.M.; Regna, K.; Muskavitch, M.A.; Poupardin, R.; Jones, C.M.; Essandoh, J.; Kétoh, G.K.; Paine, M.J.; et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014, 10, e1004236. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.L.; Wang, Z.Y.; Zou, C.S.; Cao, C.W. Transcription profiling of 12 Asian gypsy moth (Lymantria dispar) cytochrome P450 genes in response to insecticides. Arch. Insect Biochem. Physiol. 2014, 85, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.T.; Bass, C.; Williamson, M.S.; Kaussmann, M.; Wlfel, K.; Gutbrod, O.; Nauen, R. Molecular and functional characterization of CYP6BQ23, a cytochrome P450 conferring resistance to pyrethroidsin European populations of pollen beetle, Meligethes aeneus. Insect Biochem. Mol. Biol. 2014, 45, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Guitarte, J.L. Transcriptional activity of detoxification genes is altered by ultraviolet filters in Chironomus riparius. Ecotoxicol. Environ. Saf. 2018, 149, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Paz, P. Response of detoxification system genes on Chironomus riparius aquatic larvae after antibacterial agent triclosan exposures. Sci. Total Environ. 2018, 624, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.W.; Sun, L.L.; Niu, F.; Liu, P.; Chu, D.; Wang, Z.Y. Effects of phenol on metabolic activities and transcription profiles of cytochrome P450 enzymes in Chironomus kiinensis larvae. Bull. Entomol. Res. 2016, 106, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Paz, P.; Morales, M.; Martínez-Guitarte, J.L.; Morcillo, G. Characterization of a cytochrome P450 gene (CYP4G) and modulation under different exposures to xenobiotics (tributyltin, nonylphenol, bisphenol A) in Chironomus riparius aquatic larvae. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012, 155, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.M.; Park, S.Y.; Choi, J. Characterization and expression of cytochrome p450 cDNA (CYP9AT2) in Chironomus riparius fourth instar larvae exposed to multiple xenobiotics. Environ. Toxicol. Pharmacol. 2013, 36, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Yao, J.; Zhang, X.; Liu, N.; Zhu, K.Y. Comparison of gene expression profiles in the aquatic midge (Chironomus tentans) larvae exposed to two major agricultural pesticides. Chemosphere 2018, 194, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Yao, J.; Li, D.; He, Y.; Zhu, Y.C.; Zhang, X.; Zhu, K.Y. Cytochrome P450 genes from the aquatic midge Chironomus tentans: Atrazine-induced up-regulation of CtCYP6EX3 enhanced the toxicity of chlorpyrifos. Chemosphere 2017, 186, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Broza, M.; Halpern, M.; Gahanma, L.; Inbar, M. Nuisance chironomids in waste water stabilization ponds: Monitoring and action threshold assessment based on public complaints. J. Vector Ecol. 2003, 28, 31–36. [Google Scholar] [PubMed]
- Cao, C.W.; Sun, L.L.; Wen, R.R.; Li, X.P.; Wu, H.Q.; Wang, Z.Y. Toxicity and affecting factors of Bacillus thuringiensis var. israelensis on Chironomus kiiensis larvae. J. Insect Sci. 2012, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.W.; Wang, Z.Y.; Niu, C.Y.; Desneux, N.; Gao, X.W. Transcriptome profiling of Chironomus kiinensis under phenol stress using solexa sequencing technology. PLoS ONE 2013, 8, e58914. [Google Scholar] [CrossRef] [PubMed]
- Hasemann, C.A.; Kurumbail, R.G.; Boddupalli, S.S.; Peterson, J.A.; Deisenhofer, J. Structure and function of cytochrome P450: A comparative analysis of three crystal structures. Structure 1995, 3, 41–62. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect cytochrome P450. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S., Eds.; Elsevier: Oxford, UK, 2005; Volume 4, pp. 1–77. [Google Scholar]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. Identification, characterization and developmental expression of Halloween genes encoding P450 enzymes mediating ecdysone biosynthesis in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 2006, 36, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Bass, C.; Carvalho, R.A.; Oliphant, L.; Puinean, A.M.; Field, L.M.; Nauen, R.; Williamson, M.S.; Moores, G.; Gorman, K. Overexpression of a cytochrome P450 monooxygenase, CYP6ER1, is associated with resistance to imidacloprid in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2011, 20, 763–773. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Ismail, H.M.; Chandor-Proust, A.; Paine, M.J. Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120429. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Zhang, Y.X.; Yang, B.J.; Fang, J.C.; Liu, Z.W. Transcriptomic responses to different doses of cycloxaprid involved in detoxification and stress response in the whitebacked planthopper, Sogatella furcifera. Entomol. Exp. Appl. 2016, 158, 248–257. [Google Scholar] [CrossRef]
- Sun, L.L.; Wang, Z.Y.; Wu, H.Q.; Liu, P.; Zou, C.S.; Xue, X.T.; Cao, C.W. Role of ocular albinism type 1 (OA1) GPCR in Asian gypsy moth development and transcriptional expression of heat-shock protein genes. Pestic. Biochem. Physiol. 2016, 126, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Yu, Y.X.; Zhou, P.; Zhang, J.H.; Dou, L.D.; Hao, Q.; Chen, H.J.; Zhu, S.F. Identification and Knockdown of the Olfactory Receptor (OrCo) in Gypsy Moth, Lymantria dispar. Int. J. Biol. Sci. 2015, 11, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.P.; Kassa, A.; Hu, X.; Robeson, J.; McMahon, M.; Richtman, N.M.; Steimel, J.P.; Kernodle, B.M.; Crane, V.C.; Sandahl, G.; et al. Control of Western Corn Rootworm (Diabrotica virgifera virgifera) Reproduction through Plant-Mediated RNA Interference. Sci Rep. 2017, 7, 12591. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Park, Y.; Gao, X.; Zhang, X.; Yao, J.; Pang, Y.P.; Jiang, H.; Zhu, K.Y. Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci. Rep. 2012, 2, 288. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Steche, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Chu, D.; Sun, L.; Cao, C. Cytochrome P450 CYP6EV11 in Chironomus kiiensis Larvae Involved in Phenol Stress. Int. J. Mol. Sci. 2018, 19, 1119. https://doi.org/10.3390/ijms19041119
Zhang Q, Chu D, Sun L, Cao C. Cytochrome P450 CYP6EV11 in Chironomus kiiensis Larvae Involved in Phenol Stress. International Journal of Molecular Sciences. 2018; 19(4):1119. https://doi.org/10.3390/ijms19041119
Chicago/Turabian StyleZhang, Qihui, Dong Chu, Lili Sun, and Chuanwang Cao. 2018. "Cytochrome P450 CYP6EV11 in Chironomus kiiensis Larvae Involved in Phenol Stress" International Journal of Molecular Sciences 19, no. 4: 1119. https://doi.org/10.3390/ijms19041119
APA StyleZhang, Q., Chu, D., Sun, L., & Cao, C. (2018). Cytochrome P450 CYP6EV11 in Chironomus kiiensis Larvae Involved in Phenol Stress. International Journal of Molecular Sciences, 19(4), 1119. https://doi.org/10.3390/ijms19041119