Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer
Abstract
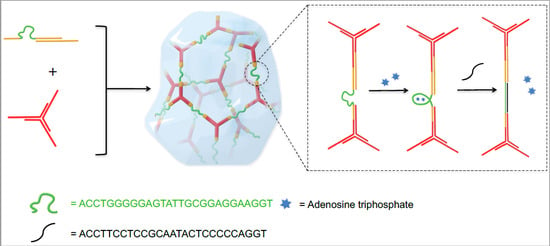
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Preparation and Characterization of DNA Assemblies
3.3. DNA Hydrogel Preparation
3.4. Rheological Characterization
3.5. Tuning Mechanical Properties of Hydrogel with ssDNA C1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Appel, E.A.; del Barrio, J.; Loh, X.J.; Scherman, O.A. Supramolecular polymeric hydrogels. Chem. Soc. Rev. 2012, 41, 6195–6214. [Google Scholar] [CrossRef] [PubMed]
- Peppas, N.A.; Hilt, J.Z.; Khademhosseini, A.; Langer, R. Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv. Mater. 2006, 18, 1345–1360. [Google Scholar] [CrossRef]
- Seliktar, D. Designing cell-compatible hydrogels for biomedical applications. Science 2012, 336, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, B.V.; Khurshid, S.S.; Fisher, O.Z.; Khademhosseini, A.; Peppas, N.A. Hydrogels in regenerative medicine. Adv. Mater. 2009, 21, 3307–3329. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Annabi, N.; Tamayol, A.; Uquillas, J.A.; Akbari, M.; Bertassoni, L.E.; Cha, C.; Camci-Unal, G.; Dokmeci, M.R.; Peppas, N.A.; Khademhosseini, A. 25th anniversary article: Rational design and applications of hydrogels in regenerative medicine. Adv. Mater. 2014, 26, 85–124. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Xing, Y.; Xi, Y.; Liu, X.; Zhou, T.; Ma, X.; Yang, Z.; Wang, S.; Liu, D. A triggered DNA hydrogel cover to envelop and release single cells. Adv. Mater. 2013, 25, 4714–4717. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Faulkner-Jones, A.; Dun, A.; Jin, J.; Chen, P.; Xing, Y.; Yang, Z.; Li, Z.; Shu, W.; Liu, D.; et al. Rapid formation of a supramolecular polypeptide-DNA hydrogel for in situ three-dimensional multilayer bioprinting. Angew. Chem. Int. Ed. Engl. 2015, 54, 3957–3961. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, C.; Cansiz, S.; Wu, C.; Xu, J.; Cui, C.; Liu, Y.; Hou, W.; Wang, Y.; Zhang, L.; et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 2015, 137, 1412–1415. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, C.; Boldt, F.; Wang, Y.; Kuan, S.; Tran, T.; Mikhalevich, V.; Förtsch, C.; Barth, H.; Yang, Z.; et al. Programmable protein-DNA hybrid hydrogels for the immobilization and release of functional proteins. Chem. Commun. 2014, 50, 14620–14622. [Google Scholar] [CrossRef] [PubMed]
- Dave, N.; Chan, M.; Huang, P.; Smith, B.; Liu, J. Regenerable DNA-functionalized hydrogels for ultrasensitive, instrument-free mercury(II) detection and removal in water. J. Am. Chem. Soc. 2010, 132, 12668–12673. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhu, Z.; Zou, Y.; Huang, Y.; Liu, D.; Jia, S.; Xu, D.; Wu, M.; Zhou, Y.; Zhou, S.; et al. Target-repsonsive “sweet” hydrogel with glucometer readout: An effective method for portable and quantitative detection of non-glucose targets. J. Am. Chem. Soc. 2013, 135, 3748–3751. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wu, C.; Liu, H.; Zou, Y.; Zhang, X.; Kang, H.; Yang, C.J.; Tan, W. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew. Chem. Int. Ed. Engl. 2010, 49, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Pan, V.; Vivek, S.; Weeks, E.; Ke, Y. Programmable DNA hydrogels assembled from multidomain DNA strands. ChemBioChem 2016, 17, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Duan, X.; Feng, Y.; Liu, L.; Wang, S.; Li, Y.; Zhu, D. Fluorescent DNA-poly(phenyl enevinylene) hybrid hydrogels for monitoring drug release. Chem. Commun. 2009, 14, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Cheng, I.; Luo, K.Q.; Mi, Y. Capture and release of protein by a reversible DNA-induced sol-gel transition system. Angew. Chem. Int. Ed. Engl. 2008, 47, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; He, K.; Zhu, R.; Liu, Z.; Zeng, S.; Huang, Y.; Nie, Z.; Yao, S. Self-assembled DNA hydrogel based on enzymatically polymerized DNA for protein encapsulation and enzyme/dnazyme hybrid cascade reaction. ACS Appl. Mater. Interfaces 2016, 8, 22801–22807. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.S.; Collins, S.D.; Anseth, K.S. Multifunctional hydrogels that promote osteogenic human mesenchymal stem cell differentiation through stimulation and sequestering of bone morphogenic protein 2. Adv. Funct. Mater. 2007, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.S.; Schwartz, M.P.; Durney, A.R.; Anseth, K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat. Mater. 2008, 7, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Kopeček, J.; Yang, J. Smart self-assembled hybrid hydrogel biomaterials. Angew. Chem. Int. Ed. Engl. 2012, 51, 7396–7417. [Google Scholar] [CrossRef] [PubMed]
- Gothelf, K.V. Lego-like DNA structures. Science 2012, 338, 1159–1160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, P.; Niu, L.; Jin, J.; Liang, D.; Li, Z.; Yang, Z.; Liu, D. pH-responsive size-tunable self-assembled DNA dendrimers. Angew. Chem. Int. Ed. Engl. 2012, 51, 11271–11274. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang, Z.; Liu, D. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014, 47, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Cheng, E.; Yang, Z. DNA-based switchable devices and materials. NPG Asia Mater. 2011, 3, 109–114. [Google Scholar] [CrossRef]
- Wilner, O.I.; Willner, I. Functionalized DNA nanostructures. Chem. Rev. 2012, 112, 2528–2556. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jia, H.; Cao, T.; Liu, D. Supramolecular hydrogels based on DNA self-assembly. Acc. Chem. Res. 2017, 50, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Jia, H.; Liu, D. pH-responsive supramolecular hydrogel based on one short strand DNA. Acta Polym. Sin. 2017, 135–142. [Google Scholar]
- Shao, Y.; Li, C.; Zhou, X.; Chen, P.; Yang, Z.; Li, Z.; Liu, D. Responsive polypeptide-DNA hydrogel crosslinked by g-quadruplex. Acta Chim. Sin. 2015, 73, 815–818. [Google Scholar]
- Cheng, E.; Xing, Y.; Chen, P.; Yang, Y.; Sun, Y.; Zhou, D.; Xu, L.; Fan, Q.; Liu, D. A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. Int. Ed. Engl. 2009, 48, 7660–7663. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Cheng, E.; Yang, Y.; Chen, P.; Zhang, T.; Sun, Y.; Yang, Z.; Liu, D. Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness. Adv. Mater. 2011, 23, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, X.; Shao, Y.; Chen, P.; Xing, Y.; Yang, Z.; Li, Z.; Liu, D. A supramolecular hydrogel with identical cross-linking point density but distinctive rheological properties. Mater. Chem. Front. 2017, 1, 654–659. [Google Scholar] [CrossRef]
- Li, C.; Chen, P.; Shao, Y.; Zhou, X.; Wu, Y.; Yang, Z.; Li, Z.; Weil, T.; Liu, D. A writable polypeptide-DNA hydrogel with rationally designed multi-modification sites. Small 2015, 11, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, C.; Shao, Y.; Chen, C.; Yang, Z.; Liu, D. Reversibly tuning mechanical properties of DNA hydrogel by DNA nanomotor. Chem. Commun. 2016, 52, 10668–10671. [Google Scholar] [CrossRef] [PubMed]
- Knowles, J.R. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980, 49, 877–919. [Google Scholar] [CrossRef] [PubMed]
- Cotrina, M.L.; Lin, J.H.; López-Garcı́a, J.C.; Naus, C.C.; Nedergaard, M. ATP-mediated glia signaling. J. Neurosci. 2000, 20, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
- Huizenga, D.E.; Szostak, J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry 1995, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sharon, E.; Freeman, R.; Liu, X.; Willner, I. Fluorescence detection of DNA, adenosine-5′-triphosphate (ATP), and telomerase activity by zinc(ІІ)-protoporphyrin IX/G-quadruplex labels. Anal. Chem. 2012, 84, 4789–4797. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Ye, D.; Zuo, X.; Li, J.; Wang, J.; Liu, H.; Hwang, M.T.; Chao, J.; Su, S.; Wang, L.; et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. 2017, 17, 5193–5198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Yang, Z.; Liu, D. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. Int. Ed. Engl. 2011, 50, 11934–11936. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Cao, T.; Xu, Y.; Dong, Y.; Liu, D. Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer. Int. J. Mol. Sci. 2018, 19, 1633. https://doi.org/10.3390/ijms19061633
Liu H, Cao T, Xu Y, Dong Y, Liu D. Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer. International Journal of Molecular Sciences. 2018; 19(6):1633. https://doi.org/10.3390/ijms19061633
Chicago/Turabian StyleLiu, Hengyuan, Tianyang Cao, Yun Xu, Yuanchen Dong, and Dongsheng Liu. 2018. "Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer" International Journal of Molecular Sciences 19, no. 6: 1633. https://doi.org/10.3390/ijms19061633
APA StyleLiu, H., Cao, T., Xu, Y., Dong, Y., & Liu, D. (2018). Tuning the Mechanical Properties of a DNA Hydrogel in Three Phases Based on ATP Aptamer. International Journal of Molecular Sciences, 19(6), 1633. https://doi.org/10.3390/ijms19061633