Nerve Growth Factor (NGF)—Receptor Survival Axis in Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Gene Expression of NGF and Protein Synthesis of NTRK1 and p75NTR in Oral Mucosa, HNSCC Tumor Tissue and in HNSCC Tumor Cell Lines
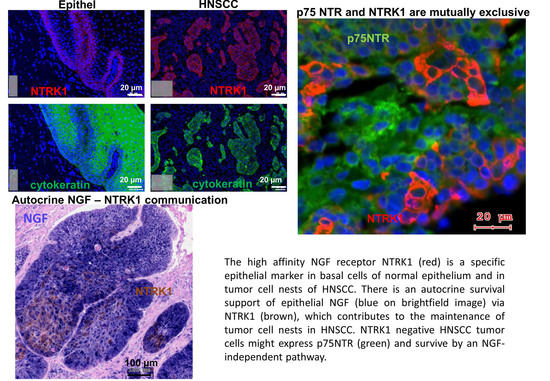
2.2. NTRK1 and p75NTR NGF Receptors Synthesis Negative Correlate in HNSCC Tumor Specimens
2.3. Patient Survival Relation of NTRK1 and p75NTR in Human Papilloma Virus (HPV) Positive and Negative HNSCC Cases
2.4. NGF Receptors in SCC-25, Detroit 562 and UPCI-SCC090 Cells
2.5. Cell Survival Effects of NGF Treatments in SCC-25, Detroit 562 and UPCI-SCC090 Cells after Cell Cycle Arrest
3. Discussion
- -
- HPV positivity is a primary factor, which is related with HNSCC patient survival benefit. NGF is expressed both in HPV+ and HPV− HNSCC, neurotrophin receptors do not show a HPV-specific distribution.
- -
- In HPV− HNSCC, high levels of both NTRK1 and p75NTR might be related with significant lower patient survival. NGF seems to be sufficiently produced by normal epithelial cells and by tumor cell nests. Most importantly the co-expression of NTRK1 and p75NTR can be observed at tumor cell nest or at whole culture level, but in fact, it means separated NTRK1- and p75NTR-positive cells.
- -
- It seems that two independent effects are needed to achieve a patient survival circumvention influence of the neurotrophin receptors in HNSCC: the first, is the NGF dependent tumor cell survival contribution of NTRK1, which is exemplified in UPCI-SCC090 cells, the second is the NGF-independent, probably p21(WAF1)-based [23] therapy resistance contribution of p75NTR, which is exemplified in Detroit 562 cells. As mentioned above, these two independent effects are also physically separated in NTRK1 and p75NTR positive cell populations.
4. Materials and Methods
4.1. Patient Samples, Immunohistochemistry
4.2. In Situ Hybridization
4.3. Image Analysis of Immunohistochemistry and In Situ Hybridization
4.4. Cell Lines
4.5. Evaluation of the NGF Receptors in Cultured Cell Lines
4.6. Cell Treatments
4.7. RNA Isolation and PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACTB | Beta-actin gene |
| BCIP | bromo-chlor-indol-phosphate |
| BDNF | brain-derived neurotrophic factor |
| DAPI | 4′,6-Diamidin-2-phenylindol |
| DIG | digoxigenin |
| EDTA | Ethylene diamine tetra acetate |
| EMT | epithelial-to-mesenchymal transition |
| HNSCC | head and neck squamous cell carcinoma |
| HPV | human papilloma virus |
| LCR | long control region |
| LNGFR | Low-affinity Nerve Growth Factor |
| MMC | mitomycin C |
| NBT | Nitro-blue-tetrazolium |
| NGF | nerve growth factor |
| NTRK | neurotrophic receptor tyrosine kinase |
| OSCC | oral squamous cell carcinoma |
| P75NTR | p75 neurotrophin receptor |
| PBS | Phosphate buffered saline |
| PCR | Polymerase chain reaction |
| SCID | severe combined immunodeficiency |
| Trk | tropomyosin-related kinase |
| UPPP | uvulopalatopharyngoplasty |
Appendix A

Appendix B
| Analyzed Cases | Investigated Question | Compared Conditions | Cases | Survival Rate, % * in up to 96 Months | Survival Time, Months (Mean ± Std. Error) | Survival Effect | p-Value by Log Rank (Mantel–Cox) Pairwise over Strata Comparison |
|---|---|---|---|---|---|---|---|
| ALL | NTRK1 level | NTRK1 level at control | 18 | 66.6% | 47.6 ± 8.77 | No significant effect | p = 0.703 |
| NTRK1 level above control | 75 | 60.0% | 52.86 ± 5.93 | ||||
| HPV-negative | NTRK1 level | NTRK1 level at control | 10 | 60.0% | 43.6 ± 11.26 | No significant effect | p = 0.997 |
| NTRK1 level above control | 54 | 57.4% | 51.58 ± 6.83 | ||||
| HPV-positive | NTRK1 level | NTRK1 level at control | 7 | 85.7% | 62.0 ± 11.63 | No significant effect | p = 0.388 |
| NTRK1 level above control | 21 | 66.7% | 41.03 ± 5.68 | ||||
| ALL | p75NTR reaction | negative | 45 | 64.4% | 47.87 ± 5.14 | No significant effect | p = 0.413 |
| positive | 48 | 58.3% | 50.39 ± 7.39 | ||||
| HPV-negative | p75NTR reaction | negative | 30 | 63.3% | 47.86 ± 6.20 | No significant effect | p = 0.279 |
| positive | 34 | 52.9% | 45.75 ± 8.50 | ||||
| HPV-positive | p75NTR reaction | negative | 14 | 71.4% | 45.22 ± 7.64 | No significant effect | p = 0.995 |
| positive | 14 | 71.4% | 50.6 ± 9.30 | ||||
| ALL | combined high NTRK1/p75NTR | NTRK1/p75NTR are NOT combined | 79 | 63.3% | 56.84 ± 5.77 | No significant effect | p = 0.139 |
| Sign. NTRK1/p75NTR combined | 14 | 50.0% | 29.25 ± 7.18 | ||||
| HPV-negative | combined high NTRK1/p75NTR | NTRK1/p75NTR are NOT combined | 54 | 61.1% | 56.41 ± 6.74 | Significant worse survival if both NTRK1 and p75NTR are high | p = 0.013 |
| Sign. NTRK1/p75NTR combined | 10 | 40.0% | 17.36 ± 7.37 | ||||
| HPV-positive | combined high NTRK1/p75NTR | NTRK1/p75NTR are NOT combined | 24 | 70.8% | 48.17 ± 7.50 | No significant effect | p = 0.552 |
| Sign. NTRK1/p75NTR combined | 4 | 75.0% | 44.50 ± 6.72 |
| Tissue Type | Number of Cases | % |
| HNSCC | 93 | 88.6 |
| UPPP epithelium | 12 | 11.4 |
| Tumor_Localization | Number of Cases | % |
| oral | 14 | 15.0 |
| nasopharynx | 4 | 4.3 |
| oropharynx | 39 | 41.9 |
| larynx | 21 | 22.6 |
| hypopharynx | 13 | 14.0 |
| other | 2 | 2.15 |
| Total | 93 | 100.0 |
| Gender | Frequency | Percent |
| Male | 85 | 90.9 |
| Female | 20 | 19.1 |
| Total | 105 | 100.0 |
| HPV | Frequency | Percent |
| Positive | 28 | 30.4 |
| Negative | 64 | 69.6 |
| Total | 92 | 100.0 |
Appendix C
Appendix C.1. TP53 mRNA Sequencing and Protein Results in SCC-25 Cells
- Query 1
- CATTTTCAGACCTATGGAAACTACTTCCTGAAAACAACGTTCTGTCCCCCTTGCCGTCCC 60||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 252
- CATTTTCAGACCTATGGAAACTACTTCCTGAAAACAACGTTCTGTCCCCCTTGCCGTCCC 311
- Query 61
- AAGCAATGGATGATTTGATGCTGTCCCCGGACGATATTGAACAATGGTTCACTGAAGACC 120||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 312
- AAGCAATGGATGATTTGATGCTGTCCCCGGACGATATTGAACAATGGTTCACTGAAGACC 371
- Query 121
- CAGGTCCAGATGAAGCTCCCAGAATGCCAGAGGCTGCTCCCCGCGTGGCCCCTGCACCAG 180|||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||
- Sbjct 372
- CAGGTCCAGATGAAGCTCCCAGAATGCCAGAGGCTGCTCCCCCCGTGGCCCCTGCACCAG 431
- Query 181
- CAGCTCCTACACCGGCGGCCCCTGCACCAGCCCCCTCCTGGCCCCTGTCATCTTCTGTCC 240||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 432
- CAGCTCCTACACCGGCGGCCCCTGCACCAGCCCCCTCCTGGCCCCTGTCATCTTCTGTCC 491
- Query 241
- CTTCCCAGAAAACCTACCAGGGCAGCTACGGTTTCCGTCTGGGCTTCTTGCATTCTGGGA 300||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 492
- CTTCCCAGAAAACCTACCAGGGCAGCTACGGTTTCCGTCTGGGCTTCTTGCATTCTGGGA 551
- Query 301
- CAGCCAAGTCTGTGACTTGCACGTACTCCCCTGCCCTCAACAAGATGTTTTGCCAACTGG 360||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 552
- CAGCCAAGTCTGTGACTTGCACGTACTCCCCTGCCCTCAACAAGATGTTTTGCCAACTGG 611
- Query 361
- CCAAGACCTGCCCTGTGCAGCTGTGGGTTGATTCCACACCCCCGCCCGGCACCCGCGTCC 420||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 612
- CCAAGACCTGCCCTGTGCAGCTGTGGGTTGATTCCACACCCCCGCCCGGCACCCGCGTCC 671
- Query 421
- GCGCCATGGCCATCTACAAGCAGTCACAGCACATGACGGAGGTTGTGAGGCGCTGCCCCC 480||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 672
- GCGCCATGGCCATCTACAAGCAGTCACAGCACATGACGGAGGTTGTGAGGCGCTGCCCCC 731
- Query 481
- ACCATGAGCGCTGCTCAGATAGCGATGGTCTGGCCCCTCCTCAGCATCTTATCCGAGTGG 540||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 732
- ACCATGAGCGCTGCTCAGATAGCGATGGTCTGGCCCCTCCTCAGCATCTTATCCGAGTGG 791
- Query 541
- AAGGAAATTTGCGTGTGGAGTATTTGGATGAC--AAACACTTTTCGACATAGTGTGGTGG 598|||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||
- Sbjct 792
- AAGGAAATTTGCGTGTGGAGTATTTGGATGACAGAAACACTTTTCGACATAGTGTGGTGG 851
- Query 599
- TGCCCTATGAGCCGCCTGAGGTTGGCTCTGACTGTACCACCATCCACTACAACTACATGT 658||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 852
- TGCCCTATGAGCCGCCTGAGGTTGGCTCTGACTGTACCACCATCCACTACAACTACATGT 911
- Query 659
- GTAACAGTTCCTGCATGGGCGGCATGAACCGGAGGCCCATCCTCACCATCATCACACTGG 718||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 912
- GTAACAGTTCCTGCATGGGCGGCATGAACCGGAGGCCCATCCTCACCATCATCACACTGG 971
- Query 719
- AAGACTCCAGTGGTAATCTACTGGGACGGAACAGCTTTGAGGTGCGTGTTTGTGCCTGTC 778||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 972
- AAGACTCCAGTGGTAATCTACTGGGACGGAACAGCTTTGAGGTGCGTGTTTGTGCCTGTC 1031
- Query 779
- CTGGGAGAGACCGGCGCACAGAGGAAGAGAATCTCCGCAAGAAAGGGGAGCCTCACCACG 838||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- Sbjct 1032
- CTGGGAGAGACCGGCGCACAGAGGAAGAGAATCTCCGCAAGAAAGGGGAGCCTCACCACG 1091
- Query 839
- AGCTGCCCCCAGGGAGCACTAAGCGAGCACTGCCCAACAACAYCAGCTCCTCTCCCCAGC 898|||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||
- Sbjct 1092
- AGCTGCCCCCAGGGAGCACTAAGCGAGCACTGCCCAACAACACCAGCTCCTCTCCCCAGC 1151
Appendix C.2. List of Changes to the Data Bank Sequence

References
- Nelson, K.B.; Grether, J.K.; Croen, L.A.; Dambrosia, J.M.; Dickens, B.F.; Jelliffe, L.L.; Hansen, R.L.; Phillips, T.M. Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann. Neurol. 2001, 49, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Duman, R.; Sanacora, G. Serum brain-derived neurotrophic factor, depression, and antidepressant medications: Meta-analyses and implications. Biol. Psychiatry 2008, 64, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.B.; Zuo, X.L.; Zhao, Q.J.; Chen, F.X.; Yang, J.; Dong, Y.Y.; Wang, P.; Li, Y.Q. Brain-derived neurotrophic factor contributes to abdominal pain in irritable bowel syndrome. Gut 2012, 61, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Kaye, D.M.; Vaddadi, G.; Gruskin, S.L.; Du, X.J.; Esler, M.D. Reduced myocardial nerve growth factor expression in human and experimental heart failure. Circ. Res. 2000, 86, E80–E84. [Google Scholar] [CrossRef] [PubMed]
- Luongo, L.; Maione, S.; Di Marzo, V. Endocannabinoids and neuropathic pain: Focus on neuron-glia and endocannabinoid-neurotrophin interactions. Eur. J. Neurosci. 2014, 39, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Hempstead, B.L.; Schleifer, L.S.; Chao, M.V. Expression of functional nerve growth factor receptors after gene transfer. Science 1989, 243, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Tebar, A.; Dechant, G.; Barde, Y.A. Binding of brain-derived neurotrophic factor to the nerve growth factor receptor. Neuron 1990, 4, 487–492. [Google Scholar] [CrossRef]
- Verge, V.M.; Merlio, J.P.; Grondin, J.; Ernfors, P.; Persson, H.; Riopelle, R.J.; Hokfelt, T.; Richardson, P.M. Colocalization of NGF binding sites, trk mRNA, and low-affinity NGF receptor mRNA in primary sensory neurons: Responses to injury and infusion of NGF. J. Neurosci. 1992, 12, 4011–4022. [Google Scholar] [CrossRef] [PubMed]
- Squinto, S.P.; Stitt, T.N.; Aldrich, T.H.; Davis, S.; Bianco, S.M.; Radziejewski, C.; Glass, D.J.; Masiakowski, P.; Furth, M.E.; Valenzuela, D.M.; et al. trkB encodes a functional receptor for brain-derived neurotrophic factor and neurotrophin-3 but not nerve growth factor. Cell 1991, 65, 885–893. [Google Scholar] [CrossRef]
- Lamballe, F.; Klein, R.; Barbacid, M. trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell 1991, 66, 967–979. [Google Scholar] [CrossRef]
- Roh, J.; Muelleman, T.; Tawfik, O.; Thomas, S.M. Perineural growth in head and neck squamous cell carcinoma: A review. Oral Oncol. 2015, 51, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, Y.K.; Wong, E.Y.; Lau, C.P.; Chan, J.P.; Wong, S.C.; Chan, A.S.; Kwan, M.P.; Tsao, S.W.; Tsang, C.M.; Lai, P.B.; et al. K252a induces anoikis-sensitization with suppression of cellular migration in Epstein-Barr virus (EBV)—Associated nasopharyngeal carcinoma cells. Investig. New Drugs 2012, 30, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Dudas, J.; Bitsche, M.; Schartinger, V.; Falkeis, C.; Sprinzl, G.M.; Riechelmann, H. Fibroblasts produce brain-derived neurotrophic factor and induce mesenchymal transition of oral tumor cells. Oral Oncol. 2011, 47, 98–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudas, J.; Fullar, A.; Bitsche, M.; Schartinger, V.; Kovalszky, I.; Sprinzl, G.M.; Riechelmann, H. Tumor-produced, active interleukin-1beta regulates gene expression in carcinoma-associated fibroblasts. Exp. Cell Res. 2011, 317, 2222–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudas, J.; Fullar, A.; Romani, A.; Pritz, C.; Kovalszky, I.; Hans Schartinger, V.; Mathias Sprinzl, G.; Riechelmann, H. Curcumin targets fibroblast-tumor cell interactions in oral squamous cell carcinoma. Exp. Cell Res. 2013, 319, 800–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kupferman, M.E.; Jiffar, T.; El-Naggar, A.; Yilmaz, T.; Zhou, G.; Xie, T.; Feng, L.; Wang, J.; Holsinger, F.C.; Yu, D.; et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene 2010, 29, 2047–2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Karatsaidis, A.; Schreurs, O.; Bjornland, T.; Sugisaki, M.; Schenck, K. NGF and its receptors TrkA and p75NTR in the epithelium of oral lichen. J. Oral Pathol. Med. 2008, 37, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Okumura, T.; Hirano, K.; Watanabe, T.; Nagata, T.; Shimada, Y.; Tsukada, K. p75 neurotrophin receptor expression is a characteristic of the mitotically quiescent cancer stem cell population present in esophageal squamous cell carcinoma. Int. J. Oncol. 2016, 48, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, R.; Mnich, K.; Gorman, A.M. Nerve growth factor (NGF)-mediated regulation of p75(NTR) expression contributes to chemotherapeutic resistance in triple negative breast cancer cells. Biochem. Biophys. Res. Commun. 2016, 478, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Marynka-Kalmani, K.; Treves, S.; Yafee, M.; Rachima, H.; Gafni, Y.; Cohen, M.A.; Pitaru, S. The lamina propria of adult human oral mucosa harbors a novel stem cell population. Stem Cells 2010, 28, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Tomellini, E.; Lagadec, C.; Polakowska, R.; Le Bourhis, X. Role of p75 neurotrophin receptor in stem cell biology: More than just a marker. Cell. Mol. Life Sci. 2014, 71, 2467–2481. [Google Scholar] [CrossRef] [PubMed]
- Soland, T.M.; Brusevold, I.J.; Koppang, H.S.; Schenck, K.; Bryne, M. Nerve growth factor receptor (p75 NTR) and pattern of invasion predict poor prognosis in oral squamous cell carcinoma. Histopathology 2008, 53, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, S.; Meignan, S.; Lagadec, C.; Germain, E.; Hondermarck, H.; Adriaenssens, E.; Le Bourhis, X. Overexpression of p75(NTR) increases survival of breast cancer cells through p21(waf1). Cell Signal. 2010, 22, 1864–1873. [Google Scholar] [CrossRef] [PubMed]
- Saleh, K.; Eid, R.; Haddad, F.G.; Khalife-Saleh, N.; Kourie, H.R. New developments in the management of head and neck cancer—Impact of pembrolizumab. Ther. Clin. Risk Manag. 2018, 14, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Gottgens, E.L.; Ostheimer, C.; Bussink, J.; Span, P.N.; Hammond, E. HPV, hypoxia and radiation response in head and neck cancer. Br. J. Radiol. 2018, 20180047. [Google Scholar] [CrossRef]
- Salazar, C.R.; Smith, R.V.; Garg, M.K.; Haigentz, M., Jr.; Schiff, B.A.; Kawachi, N.; Anayannis, N.; Belbin, T.J.; Prystowsky, M.B.; Burk, R.D.; et al. Human papillomavirus-associated head and neck squamous cell carcinoma survival: A comparison by tumor site and initial treatment. Head Neck Pathol. 2014, 8, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Gibson, L.F.; Samsell, L.; Piedimonte, G. NGF is an essential survival factor for bronchial epithelial cells during respiratory syncytial virus infection. PLoS ONE 2009, 4, e6444. [Google Scholar] [CrossRef] [PubMed]
- Kofler, B.; Borena, W.; Manzl, C.; Dudas, J.; Wegscheider, A.S.; Jansen-Durr, P.; Schartinger, V.; Riechelmann, H. Sensitivity of tumor surface brushings to detect human papilloma virus DNA in head and neck cancer. Oral Oncol. 2017, 67, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Beckett, M.A. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981, 41, 1657–1663. [Google Scholar] [PubMed]
- Chen, Y.W.; Lin, G.J.; Chia, W.T.; Lin, C.K.; Chuang, Y.P.; Sytwu, H.K. Triptolide exerts anti-tumor effect on oral cancer and KB cells in vitro and in vivo. Oral Oncol. 2009, 45, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Sinha, U.K.; Schorn, V.J.; Hochstim, C.; Chinn, S.B.; Zhu, S.; Masood, R. Increased radiation sensitivity of head and neck squamous cell carcinoma with sphingosine kinase 1 inhibition. Head Neck 2011, 33, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Steinbichler, T.B.; Metzler, V.; Pritz, C.; Riechelmann, H.; Dudas, J. Tumor-associated fibroblast-conditioned medium induces CDDP resistance in HNSCC cells. Oncotarget 2016, 7, 2508–2518. [Google Scholar] [CrossRef] [PubMed]
- Steinbichler, T.B.; Alshaimaa, A.; Maria, M.V.; Daniel, D.; Herbert, R.; Jozsef, D.; Ira-Ida, S. Epithelial-mesenchymal crosstalk induces radioresistance in HNSCC cells. Oncotarget 2018, 9, 3641–3652. [Google Scholar] [CrossRef] [PubMed]
- Sano, D.; Xie, T.X.; Ow, T.J.; Zhao, M.; Pickering, C.R.; Zhou, G.; Sandulache, V.C.; Wheeler, D.A.; Gibbs, R.A.; Caulin, C.; et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin. Cancer Res. 2011, 17, 6658–6670. [Google Scholar] [CrossRef] [PubMed]
- Peterson, W.D., Jr.; Stulberg, C.S.; Simpson, W.F. A permanent heteroploid human cell line with type B glucose-6-phosphate dehydrogenase. Proc. Soc. Exp. Biol. Med. 1971, 136, 1187–1191. [Google Scholar] [CrossRef] [PubMed]
- Peterson, W.D., Jr.; Stulberg, C.S.; Swanborg, N.K.; Robinson, A.R. Glucose-6-phosphate dehydrogenase isoenzymes in human cell cultures determined by sucrose-agar gel and cellulose acetate zymograms. Proc. Soc. Exp. Biol. Med. 1968, 128, 772–776. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Abba, M.C.; Molinolo, A.A.; Vitale-Cross, L.; Wang, Z.; Zaida, M.; Delic, N.C.; Samuels, Y.; Lyons, J.G.; Gutkind, J.S. The head and neck cancer cell oncogenome: A platform for the development of precision molecular therapies. Oncotarget 2014, 5, 8906–8923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferris, R.L.; Martinez, I.; Sirianni, N.; Wang, J.; Lopez-Albaitero, A.; Gollin, S.M.; Johnson, J.T.; Khan, S. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): A natural disease model provides insights into viral carcinogenesis. Eur. J. Cancer 2005, 41, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Bouchalova, P.; Nenutil, R.; Muller, P.; Hrstka, R.; Appleyard, M.V.; Murray, K.; Jordan, L.B.; Purdie, C.A.; Quinlan, P.; Thompson, A.M.; et al. Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status. J. Pathol. 2014, 233, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Chute, C.L.; Paulsen, M.T.; Ragland, R.L.; Howlett, N.G.; Gueranger, Q.; Glover, T.W.; Canman, C.E. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol. Cell Biol. 2010, 30, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Nicolini, V.; Fux, C.; Fussenegger, M. A novel mammalian cell-based approach for the discovery of anticancer drugs with reduced cytotoxicity on non-dividing cells. Investig. New Drugs 2004, 22, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Draper, B.K.; Komurasaki, T.; Davidson, M.K.; Nanney, L.B. Epiregulin is more potent than EGF or TGFalpha in promoting in vitro wound closure due to enhanced ERK/MAPK activation. J. Cell. Biochem. 2003, 89, 1126–1137. [Google Scholar] [CrossRef] [PubMed]
- Metzler, V.M.; Pritz, C.; Riml, A.; Romani, A.; Tuertscher, R.; Steinbichler, T.; Dejaco, D.; Riechelmann, H.; Dudas, J. Separation of cell survival, growth, migration, and mesenchymal transdifferentiation effects of fibroblast secretome on tumor cells of head and neck squamous cell carcinoma. Tumour Biol. 2017, 39, 1010428317705507. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.S.; Brody, J.R.; Dezentje, D.A.; Gallmeier, E.; Cunningham, S.C.; Swartz, M.J.; DeMarzo, A.M.; Offerhaus, G.J.; Isacoff, W.H.; Hruban, R.H.; et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin. Cancer Res. 2005, 11, 7508–7515. [Google Scholar] [CrossRef] [PubMed]
- Seaborn, T.; Ravni, A.; Au, R.; Chow, B.K.; Fournier, A.; Wurtz, O.; Vaudry, H.; Eiden, L.E.; Vaudry, D. Induction of serpinb1a by PACAP or NGF is required for PC12 cells survival after serum withdrawal. J. Neurochem. 2014, 131, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, T.; Tange, T.; Terasawa, T.; Chiba, S.; Kuwaki, T.; Miyagawa, K.; Piao, Y.F.; Miyazono, K.; Urabe, A.; Takaku, F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J. Cell Physiol. 1989, 140, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Foley, P.; Navsaria, H.A.; Sinicropi, D.; Williams-Chestnut, R.E.; Leigh, I.M. Nerve growth factor levels in cultured human skin cells: Effect of gestation and viral transformation. Neurosci. Lett. 1995, 184, 157–160. [Google Scholar] [CrossRef]
- Fox, A.J.; Patel, H.J.; Barnes, P.J.; Belvisi, M.G. Release of nerve growth factor by human pulmonary epithelial cells: Role in airway inflammatory diseases. Eur. J. Pharmacol. 2001, 424, 159–162. [Google Scholar] [CrossRef]
- Festuccia, C.; Muzi, P.; Gravina, G.L.; Millimaggi, D.; Speca, S.; Dolo, V.; Ricevuto, E.; Vicentini, C.; Bologna, M. Tyrosine kinase inhibitor CEP-701 blocks the NTRK1/NGF receptor and limits the invasive capability of prostate cancer cells in vitro. Int. J. Oncol. 2007, 30, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Bronzetti, E.; Artico, M.; Forte, F.; Pagliarella, G.; Felici, L.M.; D’Ambrosio, A.; Vespasiani, G.; Bronzetti, B. A possible role of BDNF in prostate cancer detection. Oncol. Rep. 2008, 19, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Lai, P.C.; Wu, C.C.; Hsu, S.H.; Cheng, C.C.; Lan, Y.F.; Chiu, T.H. BDNF mediated TrkB activation is a survival signal for transitional cell carcinoma cells. Int. J. Oncol. 2010, 36, 1469–1476. [Google Scholar] [PubMed]
- Dolle, L.; El Yazidi-Belkoura, I.; Adriaenssens, E.; Nurcombe, V.; Hondermarck, H. Nerve growth factor overexpression and autocrine loop in breast cancer cells. Oncogene 2003, 22, 5592–55601. [Google Scholar] [CrossRef] [PubMed]
- Dalal, R.; Djakiew, D. Molecular characterization of neurotrophin expression and the corresponding tropomyosin receptor kinases (trks) in epithelial and stromal cells of the human prostate. Mol. Cell. Endocrinol. 1997, 134, 15–22. [Google Scholar] [CrossRef]
- Johnson Chacko, L.; Blumer, M.J.F.; Pechriggl, E.; Rask-Andersen, H.; Dietl, W.; Haim, A.; Fritsch, H.; Glueckert, R.; Dudas, J.; Schrott-Fischer, A. Role of BDNF and neurotrophic receptors in human inner ear development. Cell Tissue Res. 2017, 370, 347–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.J.; Choi, Y.I.; Kim, L.; Park, I.S.; Han, J.Y.; Kim, J.M.; Chu, Y.C. Preparation of compact agarose cell blocks from the residues of liquid-based cytology samples. Korean J. Pathol. 2014, 48, 351–360. [Google Scholar] [CrossRef] [PubMed]








| Cell Lines | Untreated | NGF 100 ng/mL | Anti-NGF 100 ng/mL | Repeats | |||
|---|---|---|---|---|---|---|---|
| Trypan-Blue Negative | Trypan-Blue Positive | Trypan-Blue Negative | Trypan-Blue Positive | Trypan-Blue Negative | Trypan-Blue Positive | ||
| SCC-25 | 8.5 ± 1 | 1.7 ± 0.7 | 8.2 ± 0.6 | 1.1 ± 0.1 | 8.7 ± 0.8 | 1.2 ± 0.5 | 6 |
| Detroit 562 | 56.8 ± 23.1 | 4.6 ± 2.1 | 59.0 ± 6.7 | 2.0 ±1.1 | 56.7 ± 3.75 | 2.4 ±1.4 | 6 |
| UPCI-SCC090 | 13.9 ± 5.5 | 0.8 ± 1.0 | 19.9 ± 1.5 | 0.6 ± 0.2 | 10.2 ± 0.3 | 0.1 ± 0.1 | 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dudás, J.; Dietl, W.; Romani, A.; Reinold, S.; Glueckert, R.; Schrott-Fischer, A.; Dejaco, D.; Johnson Chacko, L.; Tuertscher, R.; Schartinger, V.H.; et al. Nerve Growth Factor (NGF)—Receptor Survival Axis in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 1771. https://doi.org/10.3390/ijms19061771
Dudás J, Dietl W, Romani A, Reinold S, Glueckert R, Schrott-Fischer A, Dejaco D, Johnson Chacko L, Tuertscher R, Schartinger VH, et al. Nerve Growth Factor (NGF)—Receptor Survival Axis in Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2018; 19(6):1771. https://doi.org/10.3390/ijms19061771
Chicago/Turabian StyleDudás, József, Wolfgang Dietl, Angela Romani, Susanne Reinold, Rudolf Glueckert, Anneliese Schrott-Fischer, Daniel Dejaco, Lejo Johnson Chacko, Raphaela Tuertscher, Volker Hans Schartinger, and et al. 2018. "Nerve Growth Factor (NGF)—Receptor Survival Axis in Head and Neck Squamous Cell Carcinoma" International Journal of Molecular Sciences 19, no. 6: 1771. https://doi.org/10.3390/ijms19061771
APA StyleDudás, J., Dietl, W., Romani, A., Reinold, S., Glueckert, R., Schrott-Fischer, A., Dejaco, D., Johnson Chacko, L., Tuertscher, R., Schartinger, V. H., & Riechelmann, H. (2018). Nerve Growth Factor (NGF)—Receptor Survival Axis in Head and Neck Squamous Cell Carcinoma. International Journal of Molecular Sciences, 19(6), 1771. https://doi.org/10.3390/ijms19061771