Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation
Abstract
:1. Introduction
2. Results
2.1. In Silico-Analysis of K+ Transporters Putatively Involved in Seed Maturation
2.2. Reverse Genetics Screen to Identify Potassium Transporter/Channel with Significant Impact on Embryo Size
2.3. Identification of Evolutionary Conserved Cis-Motifs in Target Gene Promoters
2.4. Transcriptional Regulation of Target Gene Expression during Seed Maturation
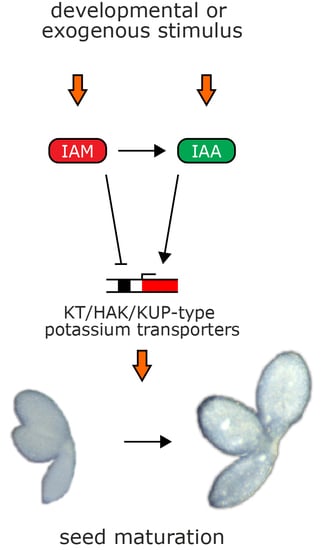
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. In Silico-Analyses
4.3. RNA Isolation and qRT-PCR
4.4. Embryo Preparation and Analysis
4.5. IAM Quantification
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cDNA | complementary DNA |
| EtOH | ethanol |
| DTT | 1,4-dithiothreitol |
| DEPC | diethyl pyrocarbonate |
| qRT-PCR | quantitative reverse transcriptase polymerase chain reaction |
| T-DNA | transfer DNA |
References
- West, M.; Harada, J.J. Embryogenesis in Higher Plants: An Overview. Plant Cell 1993, 5, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Van Wuytswinkel, O.; Castelain, M.; Bellini, C. Combined networks regulating seed maturation. Trends Plant Sci. 2007, 12, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, W.F.; Clark, J.K. Maize embryogeny: A promising experimental system. Trends Genet. 1987, 3, 3–6. [Google Scholar] [CrossRef]
- Goldberg, R.B.; Barker, S.J.; Perez-Grau, L. Regulation of gene expression during plant embryogenesis. Cell 1989, 56, 149–160. [Google Scholar] [CrossRef]
- Goldberg, R.B.; de Paiva, G.; Yadegari, R. Plant embryogenesis: Zygote to seed. Science 1994, 266, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Raz, V.; Bergervoet, J.H.; Koornneef, M. Sequential steps for developmental arrest in Arabidopsis seeds. Development 2001, 128, 243–252. [Google Scholar] [PubMed]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nambara, E.; Marion-Poll, A. ABA action and interactions in seeds. Trends Plant Sci. 2003, 8, 213–217. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicente-Carbajosa, J.; Carbonero, P. Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005, 49, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, P.J. Plant Hormones. Biosynthesis, Signal Transduction, Action! 3rd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands; Boston, MA, USA; London, UK, 2010. [Google Scholar]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jurgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Jenik, P.D.; Barton, M.K. Surge and destroy: The role of auxin in plant embryogenesis. Development 2005, 132, 3577–3585. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T.; Benkova, E.; Baurle, I.; Kientz, M.; Jurgens, G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002, 16, 1610–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardi, J.; Lanubile, A.; Li, Q.B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012, 160, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Müller, A.; Piotrowski, M.; Weiler, E.W. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 2002, 216, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schruff, M.C.; Spielman, M.; Tiwari, S.; Adams, S.; Fenby, N.; Scott, R.J. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 2006, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, C.; Wang, N.; Jiang, X.; Mao, H.; Zhu, C.; Wen, F.; Wang, X.; Lu, Z.; Yue, G.; et al. Manipulation of Auxin Response Factor 19 affects seed size in the woody perennial Jatropha curcas. Sci. Rep. 2017, 7, 40844. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Suslov, D.; Vandenbussche, F.; Kenobi, K.; Ivakov, A.; Van Der Straeten, D.; Lorences, E.P.; Mellerowicz, E.J.; Verbelen, J.P.; Vissenberg, K. Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 2013, 64, 2481–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miedes, E.; Zarra, I.; Hoson, T.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 2011, 168, 196–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Chen, H.; Ying, L.; Cai, W. AtDOF5.4/OBP4, a DOF Transcription Factor Gene that Negatively Regulates Cell Cycle Progression and Cell Expansion in Arabidopsis thaliana. Sci. Rep. 2016, 6, 27705. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.-P.; Estelle, M. Hypocotyl Transcriptome Reveals Auxin Regulation of Growth-Promoting Genes through GA-Dependent and -Independent Pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.; Ljung, K.; Fankhauser, C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 2015, 208, 198–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, R.A.; Wyn Jones, R.G. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Dreyer, I.; Uozumi, N. Potassium channels in plant cells. FEBS J. 2011, 278, 4293–4303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Porras, J.L.; Riano-Pachon, D.M.; Benito, B.; Haro, R.; Sklodowski, K.; Rodriguez-Navarro, A.; Dreyer, I. Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 2012, 3, 167. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; et al. ePlant: Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J. Arabnidopsis—An Atlas of Morphology and Development; Springer: New York, NY, USA, 1994. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Sessions, A.; Burke, E.; Presting, G.; Aux, G.; McElver, J.; Patton, D.; Dietrich, B.; Ho, P.; Bacwaden, J.; Ko, C.; et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell 2002, 14, 2985–2994. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, V.; Springer, P.; Volpe, T.; Haward, S.; Jones, J.D.; Dean, C.; Ma, H.; Martienssen, R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995, 9, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.E.; Lewis, B.D.; Spalding, E.P.; Sussman, M.R. A Role for the AKT1 Potassium Channel in Plant Nutrition. Science 1998, 280, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Dreyer, I. Structure–Function Correlates in Plant Ion Channels. In Comprehensive Biophysics; Egelman, E.H., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 234–245. [Google Scholar]
- Grabov, A. Plant KT/KUP/HAK Potassium Transporters: Single Family—Multiple FunctionsGrabov—Plant Potassium TransportersGrabov—Plant Potassium Transporters. Ann. Bot. 2007, 99, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Gajdanowicz, P.; Michard, E.; Sandmann, M.; Rocha, M.; Corrêa, L.G.G.; Ramírez-Aguilar, S.J.; Gomez-Porras, J.L.; González, W.; Thibaud, J.-B.; van Dongen, J.T.; et al. Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc. Natl. Acad. Sci. USA 2011, 108, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, I.; Gomez-Porras, J.L.; Riedelsberger, J. The potassium battery: A mobile energy source for transport processes in plant vascular tissues. New Phytol. 2017, 216, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Cuin, T.; Dreyer, I.; Michard, E. The Role of Potassium Channels in Arabidopsis thaliana Long Distance Electrical Signalling: AKT2 Modulates Tissue Excitability While GORK Shapes Action Potentials. Int. J. Mol. Sci. 2018, 19, 926. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Zhao, Y. Auxin perception and downstream events. Curr. Opin. Plant Biol. 2016, 33, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, M.; Slane, D.; Jürgens, G. Early plant embryogenesis—Dark ages or dark matter? Curr. Opin. Plant Biol. 2017, 35, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-B.; Xie, Z.-Z.; Hu, C.-G.; Zhang, J.-Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Mironova, V.V.; Omelyanchuk, N.A.; Wiebe, D.S.; Levitsky, V.G. Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genom. 2014, 15 (Suppl. 12), S4. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilfoyle, T.J.; Ulmasov, T.; Hagen, G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 1998, 54, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Liu, Z.B.; Hagen, G.; Guilfoyle, T.J. Composite structure of auxin response elements. Plant Cell 1995, 7, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Keilwagen, J.; Grau, J.; Paponov, I.A.; Posch, S.; Strickert, M.; Grosse, I. De-Novo Discovery of Differentially Abundant Transcription Factor Binding Sites Including Their Positional Preference. PLoS Comput. Biol. 2011, 7, e1001070. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; AAAI Press: Menlo Park, CA, USA, 1994. [Google Scholar]
- Frith, M.C.; Saunders, N.F.W.; Kobe, B.; Bailey, T.L. Discovering Sequence Motifs with Arbitrary Insertions and Deletions. PLoS Comput. Biol. 2008, 4, e1000071. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Costantini, E.; Landi, L.; Silvestroni, O.; Pandolfini, T.; Spena, A.; Mezzetti, B. Auxin synthesis-encoding transgene enhances grape fecundity. Plant Physiol. 2007, 143, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Mezzetti, B.; Landi, L.; Pandolfini, T.; Spena, A. The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol. 2004, 4, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molesini, B.; Rotino, G.L.; Spena, A.; Pandolfini, T. Expression profile analysis of early fruit development in iaaM-parthenocarpic tomato plants. BMC Res. Notes 2009, 2, 143. [Google Scholar] [CrossRef] [PubMed]
- Rotino, G.L.; Perri, E.; Zottini, M.; Sommer, H.; Spena, A. Genetic engineering of parthenocarpic plants. Nat. Biotechnol. 1997, 15, 1398–1401. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Parra, B.; Frerigmann, H.; Pérez Alonso, M.-M.; Carrasco Loba, V.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of Four Bifunctional Plant IAM/PAM-Amidohydrolases Capable of Contributing to Auxin Biosynthesis. Plants 2014, 3, 324–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, M.; Bai, L.; Zhuang, Z.; Fan, C.; Jiang, N.; Zhao, J.; Ma, S.; Xiang, X. Comprehensive transcriptomics and proteomics analyses of pollinated and parthenocarpic litchi (Litchi chinensis Sonn.) fruits during early development. Sci. Rep. 2017, 7, 5401. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Boij, P.; Patel, R.; Wardle, A.; Topel, M.; Jarvis, P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007, 52, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, M.; Böttcher, C.; Düchting, P.; Cheng, Y.; Zhao, Y.; Berkowitz, O.; Masle, J.; Medina, J.; Pollmann, S. The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 2013, 74, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, B.; Pilot, G.; Michard, E.; Gaymard, F.; Sentenac, H.; Thibaud, J.B. A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 2000, 12, 837–851. [Google Scholar] [PubMed]
- Marten, I.; Hoth, S.; Deeken, R.; Ache, P.; Ketchum, K.A.; Hoshi, T.; Hedrich, R. AKT3, a phloem-localized K+ channel, is blocked by protons. Proc. Natl. Acad. Sci. USA 1999, 96, 7581–7586. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, I.; Michard, E.; Lacombe, B.T.; Thibaud, J.-B. A plant Shaker-like K+ channel switches between two distinct gating modes resulting in either inward-rectifying or ‘leak’ current. FEBS Lett. 2001, 505, 233–239. [Google Scholar] [CrossRef]
- Chérel, I.; Michard, E.; Platet, N.; Mouline, K.; Alcon, C.; Sentenac, H.; Thibaud, J.-B. Physical and Functional Interaction of the Arabidopsis K+ Channel AKT2 and Phosphatase AtPP2CA. Plant Cell 2002, 14, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, M.; Skłodowski, K.; Gajdanowicz, P.; Michard, E.; Rocha, M.; Gomez-Porras, J.L.; González, W.; Corrêa, L.G.G.; Ramírez-Aguilar, S.J.; Cuin, T.A.; et al. The K+ battery-regulating Arabidopsis K+ channel AKT2 is under the control of multiple post-translational steps. Plant Signal. Behav. 2011, 6, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Michard, E.; Dreyer, I.; Lacombe, B.; Sentenac, H.; Thibaud, J.B. Inward rectification of the AKT2 channel abolished by voltage-dependent phosphorylation. Plant J. 2005, 44, 783–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michard, E.; Lacombe, B.; Porée, F.; Mueller-Roeber, B.; Sentenac, H.; Thibaud, J.-B.; Dreyer, I. A Unique Voltage Sensor Sensitizes the Potassium Channel AKT2 to Phosphoregulation. J. Gen. Physiol. 2005, 126, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, T.; Dreyer, I.; Riedelsberger, J. The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, W.H.; Wang, Y. Potassium channel AKT1 is involved in the auxin-mediated root growth inhibition in Arabidopsis response to low K+ stress. J. Integr. Plant Biol. 2017, 59, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Czempinski, K.; Frachisse, J.M.; Maurel, C.; Barbier-Brygoo, H.; Mueller-Roeber, B. Vacuolar membrane localization of the Arabidopsis ‘two-pore’ K+ channel KCO1. Plant J. 2002, 29, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Voelker, C.; Schmidt, D.; Mueller-Roeber, B.; Czempinski, K. Members of the Arabidopsis AtTPK/KCO family form homomeric vacu channels in planta. Plant J. 2006, 48, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Isner, J.C.; Begum, A.; Nuehse, T.; Hetherington, A.M.; Maathuis, F.J.M. KIN7 Kinase Regulates the Vacuolar TPK1 K+ Channel during Stomatal Closure. Curr. Biol. 2018, 28, 466–472.e4. [Google Scholar] [CrossRef] [PubMed]
- Voelker, C.; Gomez-Porras, J.L.; Becker, D.; Hamamoto, S.; Uozumi, N.; Gambale, F.; Mueller-Roeber, B.; Czempinski, K.; Dreyer, I. Roles of tandem-pore K+ channels in plants—A puzzle still to be solved. Plant Biol. 2010, 12 (Suppl. 1), 56–63. [Google Scholar] [CrossRef] [PubMed]
- Rocchetti, A.; Sharma, T.; Wulfetange, C.; Scholz-Starke, J.; Grippa, A.; Carpaneto, A.; Dreyer, I.; Vitale, A.; Czempinski, K.; Pedrazzini, E. The putative K+ channel subunit AtKCO3 forms stable dimers in Arabidopsis. Front. Plant Sci. 2012, 3, 251. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.; Janowitz, T.; Sánchez-Parra, B.; Pérez Alonso, M.-M.; Trompetter, I.; Piotrowski, M.; Pollmann, S. Arabidopsis NITRILASE 1 Contributes to the Regulation of Root Growth and Development through Modulation of Auxin Biosynthesis in Seedlings. Front. Plant Sci. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, S.; Kamiya, Y.; Nambara, E. Regulation of ABA and GA Levels During Seed Development and Germination in Arabidopsis. Annu. Plant Rev. 2007, 27. [Google Scholar] [CrossRef]
- Shuai, H.; Meng, Y.; Luo, X.; Chen, F.; Zhou, W.; Dai, Y.; Qi, Y.; Du, J.; Yang, F.; Liu, J.; et al. Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 2017, 7, 12620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, T.; Hoffmann, M.; Hentrich, M.; Pollmann, S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 2010, 89, 895–905. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Breda, A.S.; Weijers, D. Prenatal plumbing—Vascular tissue formation in the plant embryo. Physiol. Plant. 2014, 151, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Hieno, A.; Naznin, H.A.; Hyakumachi, M.; Sakurai, T.; Tokizawa, M.; Koyama, H.; Sato, N.; Nishiyama, T.; Hasebe, M.; Zimmer, A.D.; et al. ppdb: Plant promoter database version 3.0. Nucleic Acids Res. 2014, 42, D1188–D1192. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.T.; Yin, Y.; Corcoran, D.L.; Ideker, T.; Stormo, G.D.; Benos, P.V. enoLOGOS: A versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005, 33, W389–W392. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sanchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar] [CrossRef] [Green Version]
- Arvidsson, S.; Kwasniewski, M.; Riaño-Pachón, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Novak, O.; Henykova, E.; Sairanen, I.; Kowalczyk, M.; Pospisil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef] [PubMed]






| NASC Code | Name | Genotype | Gene |
|---|---|---|---|
| N662453 | hak/kt1-1 | SALK_051343C/homozygous | HAK/KT1 At2g30070 |
| N648762 | hak/kt1-2 | SALK_148762/heterozygous | HAK/KT1 At2g30070 |
| N561656 | hak/kt6-1 | SALK_061656/homozygous | HAK/KT6 At1g70300 |
| N586950 | hak/kt6-2 | SALK_086950/homozygous | HAK/KT6 At1g70300 |
| N805085 | hak/kt7-1 | SAIL_105_G04/homozygous | HAK/KT7 At5g09400 |
| N671076 | hak/kt7-2 | SALK_004133C/homozygous | HAK/KT7 At5g09400 |
| N656697 | hak/kt12-1 | SALK_083613C/homozygous | HAK/KT12 At1g60160 |
| N665909 | hak/kt12-2 | SALK_045392C/homozygous | HAK/KT12 At1g60160 |
| N670400 | kup2-1 | SALK_023287C/homozygous | KUP2 At2g40540 |
| N597636 | kup2-2 | SALK_097636/heterozygous | KUP2 At2g40540 |
| N670640 | kup4-1 | SALK_043791C/homozygous | KUP4 At4g23640 |
| N684136 | kup4-2 | SALK_071644C/homozygous | KUP4 At4g23640 |
| N670022 | kup9-1 | SALK_108080C/homozygous | KUP9 At4g19960 |
| N163575 | kup9-2 | GT_5_94315 [38] | KUP9 At4g19960 |
| N9729 | tpk1-1 | SALK_146903/homozygous | TPK1 At5g55630 |
| N661151 | tpk1-2 | SALK_131790C/homozygous | TPK1 At5g55630 |
| N662409 | tpk3-1 | SALK_049137C/homozygous | TPK3 At4g18160 |
| N663176 | tpk3-2 | SALK_085696C/homozygous | TPK3 At4g18160 |
| N684833 | kco3-1 | SALK_048607C/homozygous | KCO3 At5g46360 |
| N596038 | kco3-2 | SALK_096038/heterozygous | KCO3 At5g46360 |
| N3762 | akt1-1 | akt1-1 [39]/homozygous | AKT1 At2g26650 |
| N686273 | akt1-2 | SALK_071803C/homozygous | AKT1 At2g26650 |
| N673953 | akt2-1 | SALK_017212C/homozygous | AKT2 At4g22200 |
| N679170 | akt2-2 | SALK_141384C/homozygous | AKT2 At4g22200 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tenorio-Berrío, R.; Pérez-Alonso, M.-M.; Vicente-Carbajosa, J.; Martín-Torres, L.; Dreyer, I.; Pollmann, S. Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation. Int. J. Mol. Sci. 2018, 19, 2132. https://doi.org/10.3390/ijms19072132
Tenorio-Berrío R, Pérez-Alonso M-M, Vicente-Carbajosa J, Martín-Torres L, Dreyer I, Pollmann S. Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation. International Journal of Molecular Sciences. 2018; 19(7):2132. https://doi.org/10.3390/ijms19072132
Chicago/Turabian StyleTenorio-Berrío, Rubén, Marta-Marina Pérez-Alonso, Jesús Vicente-Carbajosa, Leticia Martín-Torres, Ingo Dreyer, and Stephan Pollmann. 2018. "Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation" International Journal of Molecular Sciences 19, no. 7: 2132. https://doi.org/10.3390/ijms19072132
APA StyleTenorio-Berrío, R., Pérez-Alonso, M. -M., Vicente-Carbajosa, J., Martín-Torres, L., Dreyer, I., & Pollmann, S. (2018). Identification of Two Auxin-Regulated Potassium Transporters Involved in Seed Maturation. International Journal of Molecular Sciences, 19(7), 2132. https://doi.org/10.3390/ijms19072132