Comparative Transcriptomics Identifies Novel Genes and Pathways Involved in Post-Traumatic Osteoarthritis Development and Progression
Abstract
:1. Introduction
2. Results
2.1. Evaluation of PTOA Development and Progression in STR/ort, C57BL/6J, and MRL/MpJ Mice Following ACL Injury
2.2. Characterizing Genes Related to Osteoarthritis Susceptibility
2.3. Early Molecular Changes Associated with PTOA Development in STR/ort, C57BL/6J, and MRL/MpJ Mice
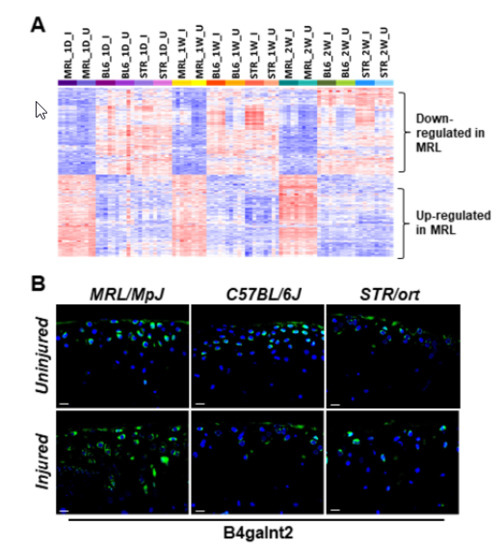
2.4. Potential Candidate Genes Associated with Enhanced Healing and Articular Cartilage Regeneration in MRL/MpJ
3. Discussion
4. Materials and Methods
4.1. Animals and Tibial Compression (TC) Joint Injury
4.2. Histological Assessment of Articular Cartilage and Joint Degeneration
4.3. Microcomputed Tomography Analysis and Osteophyte Quantification
4.4. RNA Sequencing and Data Analysis
4.5. Immunohistochemistry
4.6. Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACL | Anterior cruciate ligament |
| ECM | Extracellular matrix |
| MMP | Metalloproteinase |
| OA | Osteoarthritis |
| PTOA | Post-traumatic osteoarthritis |
| RNA-seq | RNA sequencing |
| μCT | Microcomputed tomography |
References
- Hunter, D.J.; Schofield, D.; Callander, E. The individual and socioeconomic impact of osteoarthritis. Nat. Rev. Rheumatol. 2014, 10, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Lohmander, L.S.; Englund, P.M.; Dahl, L.L.; Roos, E.M. The long-term consequence of anterior cruciate ligament and meniscus injuries: Osteoarthritis. Am. J. Sports Med. 2007, 35, 1756–1769. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.C.; Hubbard-Turner, T.; Wikstrom, E.A.; Palmieri-Smith, R.M. Epidemiology of Posttraumatic Osteoarthritis. J. Athl. Train 2017, 52, 491–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, M.T.; Felson, D.T.; Pincus, T. Analysis of the discordance between radiographic changes and knee pain in osteoarthritis of the knee. J. Rheumatol. 2000, 27, 1513–1517. [Google Scholar] [PubMed]
- Anderson, D.D.; Chubinskaya, S.; Guilak, F.; Martin, J.A.; Oegema, T.R.; Olson, S.A.; Buckwalter, J.A. Post-traumatic osteoarthritis: Improved understanding and opportunities for early intervention. J. Orthop. Res. 2011, 29, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Ramos, Y.F.; Den, H.W.; Bovée, J.V.; Bomer, N.; Van, D.B.R.; Lakenberg, N.; Keurentjes, J.C.; Goeman, J.J.; Slagboom, P.E.; Nelissen, R.G.H.H.; et al. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLoS ONE 2014, 9, e103056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, C.; Dubuc, J.E.; Montell, E.; Vergés, J.; Munaut, C.; Noël, A.; Henrotin, Y. Gene expression pattern of cells from inflamed and normal areas of osteoarthritis synovial membrane. Arthritis. Rheumatol. 2014, 66, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Klinger, P.; Beyer, C.; Ekici, A.B.; Carl, H.D.; Schett, G.; Swoboda, B.; Hennig, F.F.; Gelse, K. The Transient Chondrocyte Phenotype in Human Osteophytic Cartilage: A. Role of Pigment. Epithelium-Derived Factor? Cartilage 2013, 4, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, B.A.; Anderson, M.J.; Lee, C.A.; Williams, J.C.; Yik, J.H.; Haudenschild, D.R. Musculoskeletal changes following non-invasive knee injury using a novel mouse model of post-traumatic osteoarthritis. Osteoarthr. Cartilage 2012, 20, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Sebastian, A.; Murugesh, D.K.; Hatsell, S.; Economides, A.N.; Christiansen, B.A.; Loots, G.G. Global molecular changes in a tibial compression induced ACL rupture model of post-traumatic osteoarthritis. J. Orthop. Res. 2017, 35, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Staines, K.A.; Poulet, B.; Wentworth, D.N.; Pitsillides, A.A. The STR/ort mouse model of spontaneous osteoarthritis—An update. Osteoarthr. Cartilage 2017, 25, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Heydemann, A. The super super-healing MRL mouse strain. Front. Biol. 2012, 7, 522–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, B.D.; Furman, B.D.; Huebner, J.L.; Kraus, V.B.; Guilak, F.; Olson, S.A. Absence of posttraumatic arthritis following intraarticular fracture in the MRL/MpJ mouse. Arthrit. Rheum. 2008, 58, 744–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, J.; Rich, C.; Burkhardt, D.; Allen, J.; Herzka, A.S.; Little, C.B. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthr. Cartilage 2008, 16, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Osterberg, A.; Thiem, D.; Herlyn, P.; Mittlmeier, T.; Frerich, B.; Müller-Hilke, B. Subchondral bone sclerosis and cancellous bone loss following OA induction depend on the underlying bone phenotype. Jt. Bone Spine 2017, 84, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Leferovich, J.; Zhang, X.M.; Bedelbaeva, K.; Gourevitch, D.; Hatcher, C.J.; Basson, C.T.; Heber-Katz, E.; Marx, K.A. Keratin gene expression profiles after digit amputation in C57BL/6 vs. regenerative MRL mice imply an early regenerative keratinocyte activated-like state. Physiol. Genom. 2013, 45, 409–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Singhal, N.; Serinagaoglu, Y.; Chandrasekharan, K.; Joshi, M.; Bauer, J.A.; Janssen, P.M.L.; Martin, P.T. Deletion of Galgt2 (B4Galnt2) reduces muscle growth in response to acute injury and increases muscle inflammation and pathology in dystrophin-deficient mice. Am. J. Pathol. 2015, 185, 2668–2684. [Google Scholar] [CrossRef] [PubMed]
- Pasold, J.; Engelmann, R.; Keller, J.; Joost, S.; Marshall, R.P.; Frerich, B.; Müller-Hilke, B. High bone mass in the STR/ort mouse results from increased bone formation and impaired bone resorption and is associated with extramedullary hematopoiesis. J. Bone Min. Metab. 2013, 31, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Sokolove, J.; Lepus, C.M. Role of inflammation in the pathogenesis of osteoarthritis: Latest findings and interpretations. Ther. Adv. Musculoskelet. Dis. 2013, 5, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Kyostio-Moore, S.; Nambiar, B.; Hutto, E.; Ewing, P.J.; Piraino, S.; Berthelette, P.; Sookdeo, C.; Matthews, G.; Armentano, D. STR/ort mice, a model for spontaneous osteoarthritis, exhibit elevated levels of both local and systemic inflammatory markers. Comp. Med. 2011, 61, 346–355. [Google Scholar] [PubMed]
- Lewis, J.S., Jr.; Furman, B.D.; Zeitler, E.; Huebner, J.L.; Kraus, V.B.; Guilak, F.; Olson, S.A. Genetic and cellular evidence of decreased inflammation associated with reduced incidence of posttraumatic arthritis in MRL/MpJ mice. Arthritis. Rheum. 2013, 65, 660–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heber-Katz, E.; Naviaux, R.K. The MRL Mouse: A Model of Regeneration and Cancer. In Murine Models, Energy Balance, and Cancer; Berger, N.A., Ed.; Springer International Publishing: Cham, Switzerland, 2015; Volume 10, pp. 47–64. [Google Scholar]
- Gourevitch, D.; Clark, L.; Chen, P.; Seitz, A.; Samulewicz, S.J.; Heber-Katz, E. Matrix metalloproteinase activity correlates with blastema formation in the regenerating MRL mouse ear hole model. Dev. Dyn. 2003, 226, 377–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, B.; Klassen, H.; Yang, L.; Chen, D.F.; Young, M.J. Elevated MMP Expression in the MRL Mouse Retina Creates a Permissive Environment for Retinal Regeneration. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1686–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrage, P.S.; Mix, K.S.; Brinckerhoff, C.E. Matrix metalloproteinases: Role in arthritis. Front. Biosci. 2006, 11, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flannelly, J.; Chambers, M.G.; Dudhia, J.; Hembry, R.M.; Murphy, J.; Mason, R.M.; Bayliss, M.T. Metalloproteinase and tissue inhibitor of metalloproteinase expression in the murine STR/ort model of osteoarthritis. Osteoarthr. Cartilage 2002, 10, 722–733. [Google Scholar] [CrossRef]
- Almodovar-Garcia, K.; Kwon, M.; Samaras, S.E.; Davidson, J.M. ANKRD1 acts as a transcriptional repressor of MMP13 via the AP-1 site. Mol. Cell Biol. 2014, 34, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- Samaras, S.E.; Almodóvar-García, K.; Wu, N.; Yu, F.; Davidson, J.M. Global deletion of Ankrd1 results in a wound-healing phenotype associated with dermal fibroblast dysfunction. Am. J. Pathol. 2015, 185, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Bauman, W.A.; Cardozo, C. ANKRD1 modulates inflammatory responses in C2C12 myoblasts through feedback inhibition of NF-kappaB signaling activity. Biochem. Biophys. Res. Commun. 2015, 464, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.; Ritchie, G.R.S.; Roumeliotis, T.I.; Jayasuriya, R.L.; Clark, M.J.; Brooks, R.A.; Binch, A.L.A.; Shah, K.M.; Coyle, R.; Pardo, M.; et al. Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis. Sci. Rep. 2017, 7, 8935. [Google Scholar] [CrossRef] [PubMed]
- Bouxsein, M.L.; Boyd, S.K.; Christiansen, B.A.; Guldberg, R.E.; Jepsen, K.J.; Müller, R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Min. Res. 2010, 25, 1468–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.C.; Christiansen, B.A.; Murugesh, D.K.; Sebastian, A.; Hum, N.R.; Collette, N.M.; Hatsell, S.; Economides, A.N.; Blanchette, C.D.; Loots, G.G. SOST/Sclerostin Improves Post Traumatic Osteoarthritis and Inhibits MMP2/3 Expression After Injury. J. Bone Min. Res. 2018, 33, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]






| MRL | BL6 | STR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | 1D | 1W | 2W | 1D | 1W | 2W | 1D | 1W | 2W |
| Ccl2 | 3.61 | ns | ns | 3.23 | ns | ns | 3.53 | 1.26 | 1.14 |
| Ccl7 | 4.52 | 2.27 | ns | 3.73 | 1.97 | ns | 4.21 | 1.58 | 2.02 |
| Ccl8 | 2.52 | 2.9 | ns | 1.66 | 1.81 | ns | 1.04 | 1.23 | 1.84 |
| Ccl17 | ns | ns | ns | ns | ns | ns | ns | ns | 1.99 |
| Ccl19 | ns | ns | ns | 0.67 | ns | ns | ns | ns | ns |
| Ccl20 | ns | ns | ns | 3.24 | ns | ns | 4.73 | 4.27 | ns |
| Ccl28 | ns | ns | ns | 0.72 | ns | ns | ns | ns | ns |
| Csf1 | 0.69 | ns | ns | ns | ns | ns | 0.59 | ns | ns |
| Cxcl1 | 2.8 | ns | ns | 2.59 | ns | ns | 3.02 | 1.51 | 1.87 |
| Cxcl2 | ns | ns | ns | ns | ns | ns | 2.86 | 2.13 | ns |
| Cxcl3 | ns | ns | ns | 3.23 | ns | ns | 4.17 | 1.81 | ns |
| Cxcl5 | 1.53 | ns | ns | 2.48 | ns | ns | 3.06 | 1.18 | ns |
| Cxcl14 | 1.52 | 0.59 | ns | 0.97 | ns | ns | 1.87 | ns | 1.18 |
| Cxcl16 | 1.06 | 0.91 | ns | 1.72 | 0.88 | ns | 0.83 | 1.17 | 0.78 |
| Il1b | ns | ns | ns | ns | ns | ns | 0.64 | ns | ns |
| Il5 | ns | ns | ns | ns | 2.03 | ns | ns | ns | ns |
| Il6 | 2.29 | ns | ns | 2.29 | ns | ns | 3.24 | 1.73 | 1.95 |
| Il11 | 2.7 | 1.68 | ns | 2.82 | 1.67 | 2.02 | 4.01 | 2.28 | 2.82 |
| Il17d | ns | 1.15 | ns | ns | 1.01 | ns | ns | ns | ns |
| Il33 | 1.17 | 1.5 | ns | 1.71 | 1.06 | 1.2 | 1.28 | 1.58 | 1.25 |
| Lif | 1.16 | ns | ns | ns | ns | ns | 1.16 | ns | ns |
| Tnf | ns | ns | ns | ns | ns | ns | 0.77 | ns | ns |
| Tnfsf9 | ns | ns | ns | 1.27 | ns | ns | ns | ns | ns |
| Tnfsf15 | ns | ns | ns | ns | 1.52 | ns | ns | 1.04 | ns |
| Tnfsf18 | ns | ns | ns | ns | ns | ns | 1.58 | 2.26 | 1.52 |
| MRL | BL6 | STR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | 1D | 1W | 2W | 1D | 1W | 2W | 1D | 1W | 2W |
| Adam5 | ns | ns | ns | ns | ns | ns | ns | 0.78 | ns |
| Adam9 | ns | ns | ns | 0.96 | ns | ns | ns | 0.66 | ns |
| Adam12 | 0.65 | 0.73 | ns | 0.96 | 1.11 | ns | 0.72 | 1.6 | 0.73 |
| Adam23 | ns | 1.1 | 0.96 | 1.01 | 1.04 | 1.13 | ns | 1.55 | 1.66 |
| Adamts1 | 0.71 | ns | ns | 1.02 | ns | ns | 1.31 | 1.01 | ns |
| Adamts3 | ns | ns | ns | ns | 0.83 | ns | ns | 0.99 | 0.73 |
| Adamts4 | 1.81 | 1.21 | ns | 1.66 | 1.56 | ns | 1.64 | 1.73 | 1.43 |
| Adamts6 | ns | 0.67 | ns | ns | 0.97 | ns | ns | 0.63 | 0.77 |
| Adamts7 | ns | ns | ns | ns | 0.71 | ns | ns | 0.66 | 0.94 |
| Adamts8 | 1.27 | ns | ns | ns | ns | ns | ns | ns | ns |
| Adamts12 | ns | 1.47 | ns | 1.02 | 1.61 | 0.96 | ns | 1.6 | 1.11 |
| Adamts14 | ns | ns | ns | ns | 0.64 | ns | ns | 0.71 | ns |
| Adamts15 | ns | 1.31 | 0.84 | ns | 0.97 | 0.79 | ns | 0.64 | 1.35 |
| Adamts16 | ns | 3.05 | 2.6 | 1.08 | 3.75 | 3.37 | ns | 1.52 | 3.35 |
| Adamts17 | ns | ns | ns | ns | ns | 0.7 | ns | ns | ns |
| Adamtsl1 | ns | 0.84 | ns | 0.86 | 0.99 | 0.62 | 0.62 | 1.33 | 0.98 |
| Adamtsl2 | 1.48 | ns | ns | ns | ns | ns | 0.86 | ns | ns |
| Adamtsl3 | 0.87 | 1.11 | ns | 1.37 | 0.95 | ns | ns | 1.25 | 0.59 |
| Adamtsl4 | ns | 1.01 | ns | 0.61 | 0.68 | ns | ns | ns | ns |
| Aebp1 | ns | 1.54 | 0.88 | 1 | 1.41 | 0.84 | ns | 1.54 | 1.16 |
| Agbl2 | ns | 1.38 | ns | 1.41 | 1.73 | ns | ns | 1.69 | ns |
| Anpep | ns | 1.44 | 0.75 | 1.32 | 1.58 | 1.08 | ns | 1.18 | 1.44 |
| Cpxm1 | ns | 1.27 | 0.77 | ns | 1.47 | 0.85 | ns | 1.35 | 0.82 |
| Cpxm2 | ns | 1.39 | 0.91 | ns | 1.31 | 1.1 | ns | 1.43 | 1.36 |
| Dpep2 | 1.51 | 0.83 | ns | 1.61 | 0.7 | ns | 1.03 | 0.62 | 0.95 |
| Mmp2 | ns | 1.36 | 1.02 | ns | 1.67 | 1.15 | ns | 1.31 | 1.42 |
| Mmp3 | 2.39 | 2.88 | 2.42 | 1.99 | 2.36 | 2.12 | 1.97 | ns | 2.57 |
| Mmp11 | ns | ns | ns | ns | ns | ns | ns | 0.83 | ns |
| Mmp12 | ns | ns | 1.39 | 1.62 | 1.26 | 1.51 | ns | 1.44 | 1.24 |
| Mmp14 | ns | 0.91 | ns | ns | 1.26 | 0.66 | ns | 1.51 | 1.08 |
| Mmp19 | 1.22 | 1.42 | 0.73 | 1.4 | 1.12 | 1.01 | 0.98 | 0.92 | 1.69 |
| Naalad2 | ns | ns | ns | ns | ns | ns | ns | 0.66 | ns |
| Pappa2 | 0.8 | ns | ns | ns | 0.66 | 0.83 | ns | ns | 0.85 |
| Tll1 | ns | 0.87 | ns | 0.95 | 1.12 | 1.01 | 0.87 | 1.41 | 0.96 |
| Trabd2b | ns | 0.61 | ns | 0.64 | 0.97 | 1.04 | ns | ns | 0.81 |
| Gene Symbol | Gene Name |
|---|---|
| Genes with higher expression in MRL/MpJ compared to C57BL/6J and STR/ort | |
| Tpsab1 | tryptase alpha/beta 1 |
| Ccdc38 | coiled-coil domain containing 38 |
| Aox4 | Aldehyde oxidase 4 |
| B4galnt2 | Beta-1,4-N-acetyl-galactosaminyl transferase 2 |
| Vwa5a | von Willebrand factor A domain containing 5A |
| Genes with lower expression in MRL/MpJ compared to C57BL/6J and STR/ort | |
| Trim12a | tripartite motif-containing 12A |
| Mamdc2 | MAM domain containing 2 |
| Serpina3b | serine (or cysteine) peptidase inhibitor, clade A, member 3B |
| Rab6b | RAB6B, member RAS oncogene family |
| Capg | capping protein (actin filament), gelsolin-like |
| Myoc | Myocilin |
| Fam171b | family with sequence similarity 171, member B |
| H2-D1 | histocompatibility 2, D region locus 1 |
| Slc15a2 | solute carrier family 15 (H+/peptide transporter), member 2 |
| Ccdc109b | Coiled-coil domain containing 109B |
| Thnsl2 | Threonine synthase-like 2 |
| Pccb | propionyl Coenzyme A carboxylase, beta polypeptide |
| Gpx3 | glutathione peroxidase 3 |
| Ezh1 | enhancer of zeste 1 polycomb repressive complex 2 subunit |
| Acsf2 | Acyl-CoA synthetase family member 2 |
| Pycard | PYD and CARD domain containing |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sebastian, A.; Chang, J.C.; Mendez, M.E.; Murugesh, D.K.; Hatsell, S.; Economides, A.N.; Christiansen, B.A.; Loots, G.G. Comparative Transcriptomics Identifies Novel Genes and Pathways Involved in Post-Traumatic Osteoarthritis Development and Progression. Int. J. Mol. Sci. 2018, 19, 2657. https://doi.org/10.3390/ijms19092657
Sebastian A, Chang JC, Mendez ME, Murugesh DK, Hatsell S, Economides AN, Christiansen BA, Loots GG. Comparative Transcriptomics Identifies Novel Genes and Pathways Involved in Post-Traumatic Osteoarthritis Development and Progression. International Journal of Molecular Sciences. 2018; 19(9):2657. https://doi.org/10.3390/ijms19092657
Chicago/Turabian StyleSebastian, Aimy, Jiun C. Chang, Melanie E. Mendez, Deepa K. Murugesh, Sarah Hatsell, Aris N. Economides, Blaine A. Christiansen, and Gabriela G. Loots. 2018. "Comparative Transcriptomics Identifies Novel Genes and Pathways Involved in Post-Traumatic Osteoarthritis Development and Progression" International Journal of Molecular Sciences 19, no. 9: 2657. https://doi.org/10.3390/ijms19092657
APA StyleSebastian, A., Chang, J. C., Mendez, M. E., Murugesh, D. K., Hatsell, S., Economides, A. N., Christiansen, B. A., & Loots, G. G. (2018). Comparative Transcriptomics Identifies Novel Genes and Pathways Involved in Post-Traumatic Osteoarthritis Development and Progression. International Journal of Molecular Sciences, 19(9), 2657. https://doi.org/10.3390/ijms19092657