CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L.
Abstract
:1. Introduction
2. Results
2.1. Sequence Identification and Expression Analysis of BnWRKY11 and BnWRKY70 Genes in B. napus
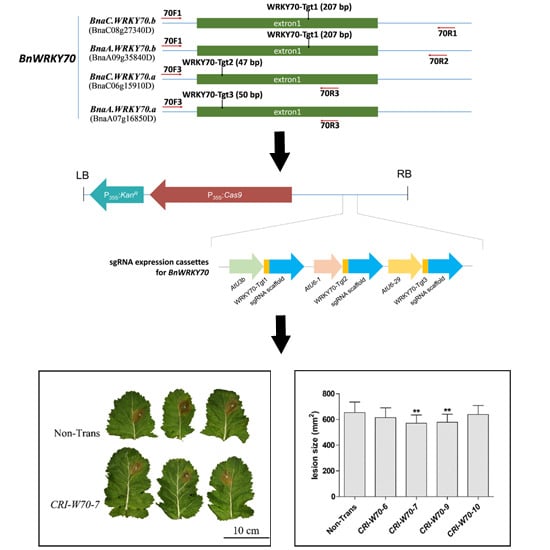
2.2. CRISPR/Cas9 Binary Vector Construction, Rapeseed Transformation and Screening of Positive Transformants
2.3. Confirmation of Cas9-Induced Mutagenesis in Transgenic Plants of B. napus
2.4. Variety and Frequency of Mutations
2.5. Mutagenesis in T1 Plants
2.6. BnWRKY70 Mutants Enhance Resistance to S. sclerotiorum
3. Discussion
4. Materials and Methods
4.1. Target Design and Vector Construction for Targeted Gene Mutation
4.2. Genetic Transformation of B. napus
4.3. Mutation Analysis
4.4. S. sclerotiorum Infection Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
Abbreviations
| CRISPR | Clustered, regularly interspaced, palindromic repeat |
| sgRNA | Single guide RNA |
| PAM | Protospacer adjacent motif |
| ZFN | Zinc finger nucleases |
| TALEN | Transcription activator-like effector nucleases |
| TFs | Transcription factors |
| hpi | hours post-inoculation |
| AS | Acetosyringone |
| Cef | Cefotaxime |
| CTAB | Hexadecyl trimethyl ammonium brom |
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J. A CRISPR view of development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barakate, A.; Stephens, J. An overview of CRISPR-based tools and their improvements: New opportunities in understanding plant-pathogen interactions for better crop protection. Front. Plant Sci. 2016, 7, 765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Malina, A.; Mills, J.R.; Cencic, R.; Yan, Y.F.; Fraser, J.; Schippers, L.M.; Paquet, M.; Dostie, J.; Pelletier, J. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013, 27, 2602–2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.Z.; Lu, G.Q.; Xie, Z.; Lou, M.L.; Luo, J.; Guo, L.; Zhang, Y. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014, 24, 1009–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, T.T.; Zhou, N.; Huang, J.; Koirala, P.; Xu, M.; Fung, R.; Wu, F.; Mo, Y.Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2014, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Minkenberg, B.; Xie, K.B.; Yang, Y.N. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 2017, 89, 636–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arazoe, T.; Miyoshi, K.; Yamato, T.; Ogawa, T.; Ohsato, S.; Arie, T.; Kuwata, S. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol. Bioeng. 2015, 112, 2543–2549. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.L.; Sun, B.B.; Yang, J.J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 System. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Z.; Yang, B.; Weeks, D.P. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 2014, 9, e99225. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Wang, G.H.; Ma, S.Y.; Xie, X.D.; Wu, X.W.; Zhang, X.T.; Wu, Y.Q.; Zhao, P.; Xia, Q.Y. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Fish, M.B.; Fisher, M.; Oomen-Hajagos, J.; Thomsen, G.H.; Grainger, R.M. Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. Genesis 2013, 51, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S.; et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.; Wang, Z.X.; Hu, Y.Y.; Wu, Y.D.; Luo, Z.; Yang, Z.P.; Zu, Y.; Li, W.Y.; Huang, P.; Tong, X.J.; et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013, 41, e141. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.B.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.Z.; Zhou, H.H.; Bi, H.H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.Y.; Mao, Y.F.; Xu, N.F.; Zhang, B.T.; Wei, P.L.; Yang, D.L.; Wang, Z.; Zhang, Z.J.; Zheng, R.Z.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Hahn, F.; Mantegazza, O.; Greiner, A.; Hegemann, P.; Eisenhut, M.; Weber, A.P. An efficient visual screen for CRISPR/Cas9 activity in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.W.; Wang, Y.P.; Li, J.; Gao, C.X. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.-A. Production of low-Cs+ rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR-Cas system. Plant J. 2017, 91, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.P.; Liu, J.X.; Chen, K.L.; Qiu, J.L.; Gao, C.X. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.D.; Ostergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [PubMed]
- Braatz, J.; Harloff, H.J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wu, J.J.; Tang, T.; Liu, K.D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, K.Y.; Li, H.L.; Han, S.Q.; Meng, Q.W.; Khan, S.U.; Fan, C.C.; Xie, K.B.; Zhou, Y.M. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018, 16, 1322–1355. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hao, M.Y.; Wang, W.X.; Wang, H.; Chen, F.; Chu, W.; Zhang, B.H.; Mei, D.S.; Cheng, H.T.; Hu, Q. An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 2018, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Saharan, G.S.; Mehta, N. Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management; Springer: Heidelberg, Germany, 2008; pp. 1–485. [Google Scholar]
- Wu, J.; Zhao, Q.; Liu, S.; Shahid, M.; Lan, L.; Cai, G.Q.; Zhang, C.Y.; Fan, C.C.; Wang, Y.P.; Zhou, Y.M. Genome-wide association study identifies new loci for resistance to Sclerotinia stem rot in Brassica napus. Front. Plant Sci. 2016, 7, 1418. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.G.; Zhang, L.P.; Li, D.B.; Wang, F.; Yu, D.Q. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Journot-Catalino, N.; Somssich, I.E.; Roby, D.; Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 2006, 18, 3289–3302. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.H.; Meng, X.Z.; Liu, Y.D.; Zheng, Z.Y.; Chen, Z.X.; Zhang, S.Q. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Sham, A.; Moustafa, K.; Al-Shamisi, S.; Alyan, S.; Iratni, R.; AbuQamar, S. Microarray analysis of Arabidopsis WRKY33 mutants in response to the necrotrophic fungus Botrytis cinerea. PLoS ONE 2017, 12, e0172343. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.C.; Lai, Z.B.; Fan, B.F.; Chen, Z.X. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.H.; Eschen-Lippold, L.; Pecher, P.; Hoehenwarter, W.; Sinha, A.K.; Scheel, D.; Lee, J. Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 61. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.R.; Dong, Q.Y.; Yu, D.Q. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185–186, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.H.; Huang, Z.Y.; Xie, P.; Gu, C.; Li, K.; Wang, D.C.; Yu, Y.Y.; Fan, Z.H.; Wang, C.J.; Wang, Y.P.; et al. Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 2016, 67, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fang, H.D.; Chen, Y.; Chen, K.P.; Li, G.Y.; Gu, S.L.; Tan, X.L. Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol. Plant Pathol. 2014, 15, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.A.; Kracher, B.; Ziegler, J.; Birkenbihl, R.P.; Somssich, I.E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 2015, 4, e07295. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, X.X.; Wang, M.R.; Wen, J.; Yi, B.; Shen, J.X.; Ma, C.Z.; Fu, T.D.; Tu, J.X. Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 2018, 16, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Raman, R.; Coombes, N.; Song, J.; Diffey, S.; Kilian, A.; Lindbeck, K.; Barbulescu, D.M.; Batley, J.; Edwards, D.; et al. Genome-wide association study identifies new loci for resistance to leptosphaeria maculans in canola. Front. Plant Sci. 2016, 7, 1513. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, Q.; Yang, Q.Y.; Liu, H.; Li, Q.Y.; Yi, X.Q.; Cheng, Y.; Guo, L.; Fan, C.C.; Zhou, Y.M. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 2016, 6, 19007. [Google Scholar] [CrossRef] [PubMed]
- Initiative, A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, L.; King, G.J. Standardized gene nomenclature for the Brassica genus. Plant Methods 2008, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. Function genomics of abiotic stress tolerance in plants: A CRISPR approach. Front. Plant Sci. 2015, 6, 375. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhang, H.M.; Lou, D.J.; Yu, D.Q. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 2016, 6, 21451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.J.; Song, N.; Sun, S.L.; Yang, W.L.; Zhao, H.M.; Song, W.B.; Lai, J.S. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genomics 2016, 43, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Zhang, Q.Y.; Zhu, Q.L.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.F.; Li, H.Y.; Lin, Y.R.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Munoz-Blanco, J.; Caballero, J.L. Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J. Exp. Bot. 2009, 60, 3043–3065. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.B.; Wang, F.; Zheng, Z.Y.; Fan, B.F.; Chen, Z.X. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulker, B.; Shahid Mukhtar, M.; Somssich, I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Nolan, T.; Ye, H.X.; Zhang, M.C.; Tong, H.N.; Xin, P.Y.; Chu, J.F.; Chu, C.C.; Li, Z.H.; Yin, Y.H. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, C.M.; Zhu, Q.; Gao, B.D.; Ke, S.Y.; Yu, W.C.; Xie, D.X.; Peng, W. Transcription factor WRKY70 displays important but no indispensable roles in jasmonate and salicylic acid signaling. J. Integr. Plant Biol. 2008, 50, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Lu, Y.; Bethke, G.; Harrison, B.T.; Hatsugai, N.; Katagiri, F.; Glazebrook, J. WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. N. Phytol. 2017, 217, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Piatek, A.; Stewart, C.N., Jr. Genome engineering via TALENs and CRISPR/Cas9 systems: Challenges and perspectives. Plant Biotechnol. J. 2014, 12, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Yuan, J.; Wang, R.; Liu, Y.; Birchler, J.A.; Han, F.P. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics 2016, 43, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, D.S.; Zhang, J.Z.; Huang, Q.P.; Qin, G.J.; Zhang, X.; Wan, J.M.; Gu, H.Y.; Qu, L.J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rietz, S.; Bernsdorff, F.E.; Cai, D.G. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 2012, 63, 5507–5519. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [PubMed]
- De Block, M.; De Brouwer, D.; Tenning, P. Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol. 1989, 91, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cai, G.Q.; Tu, J.Y.; Li, L.X.; Liu, S.; Luo, X.P.; Zhou, L.P.; Fan, C.C.; Zhou, Y.M. Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS ONE 2013, 8, e67740. [Google Scholar] [CrossRef] [PubMed]






| Target Gene (Number of Transformants) | Copies | Target | Amplification Primer | No. of Plants with Mutations | Mutation Frequency (%) |
|---|---|---|---|---|---|
| BnWRKY1122 | BnaA.WRKY11.a | WRKY11-Tgt1, WRKY11-Tgt3 | 11subF/11subR→11F/11R | 0, 7 | 0, 31.8% |
| BnaC.WRKY11.a | WRKY11-Tgt2, WRKY11-Tgt3 | 11F/11R | 12, 9 | 54.5%, 40.9% | |
| BnWRKY708 | BnaC.WRKY70.b | WRKY70-Tgt1 | 70F3/70R3 | 0 | 0 |
| BnaA.WRKY70.b | WRKY70-Tgt1 | 70F3/70R3 | 3 | 37.5% | |
| BnaC.WRKY70.a | WRKY70-Tgt2 | 70F1/70R2 | 0 | 0 | |
| BnaA.WRKY70.a | WRKY70-Tgt3 | 70F1/70R1 | 3 | 37.5% |
| Line | Number of Examined Plants | Cas9:sgRNA Constructs a | Number of Edited Plants b | |||
|---|---|---|---|---|---|---|
| BnWRKY11 | ||||||
| BnaA.WRKY11.a (WRKY11-Tgt1) | BnaA.WRKY11.a (WRKY11-Tgt3) | BnaC.WRKY11.a (WRKY11-Tgt2) | BnaC.WRKY11.a (WRKY11-Tgt3) | |||
| CRI-W11-6 | 10 | 10 | 10 | 10 | 0 | 10 |
| CRI-W11-10 | 10 | 10 | 10 | 10 | 0 | 10 |
| BnWRKY70 | ||||||
| BnaA.WRKY70.b | BnaA.WRKY70.a | BnaC.WRKY70.a | BnaC.WRKY70.b | |||
| CRI-W70-6 | 10 | 8 | 8 | 8 | 0 | 8 |
| CRI-W70-7 | 10 | 8 | 8 | 8 | 0 | 8 |
| CRI-W70-9 | 10 | 10 | 10 | 10 | 0 | 10 |
| CRI-W70-10 | 10 | 8 | 10 | 10 | 0 | 10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2716. https://doi.org/10.3390/ijms19092716
Sun Q, Lin L, Liu D, Wu D, Fang Y, Wu J, Wang Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. International Journal of Molecular Sciences. 2018; 19(9):2716. https://doi.org/10.3390/ijms19092716
Chicago/Turabian StyleSun, Qinfu, Li Lin, Dongxiao Liu, Dewei Wu, Yujie Fang, Jian Wu, and Youping Wang. 2018. "CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L." International Journal of Molecular Sciences 19, no. 9: 2716. https://doi.org/10.3390/ijms19092716
APA StyleSun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., & Wang, Y. (2018). CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. International Journal of Molecular Sciences, 19(9), 2716. https://doi.org/10.3390/ijms19092716