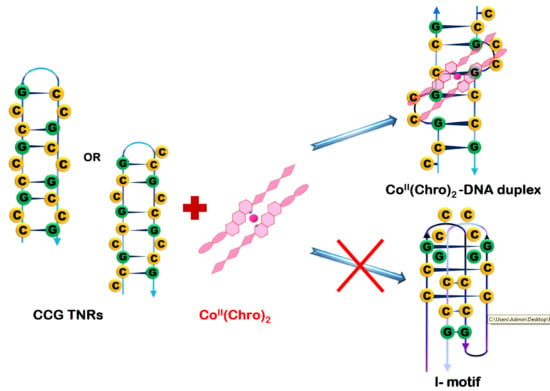
CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Abstract
:1. Introduction
2. Results
2.1. A Non-Canonical DNA Structure of the dT(CCG)3A Sequence Contains an i-Motif Tetraplex Core
2.2. Stabilization of the i-Motif Tetraplex by Water Hydration
2.3. CoII(Chro)2 Complex Induces Conformational Changes in the d[T(CCG)3A]2 DNA Duplex
2.4. Stabilization of the CoII(Chro)2-d[T(CCG)3A]2 Complex by Interacting with Cobalt(II) Ions and Water Hydration
2.5. The Cobalt–Chro Complex Specifically Recognizes the Hairpin Structure of CCG TNRs
3. Discussion
4. Materials and Methods
4.1. Chemicals and Oligonucleotides
4.2. Melting Temperature Measurements
4.3. SPR Analysis
4.4. Crystallization of d(T(CCG)3A) and CoII(Chro)2-d[T(CCG)3A]2 Complex
4.5. Data Collection, Processing, and Refinement of d(T(CCG)3A) and CoII(Chro)2-d[T(CCG)3A]2 Complex Structures
4.6. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TNR | Trinucleotide repeats |
| FMR1 | Fragile X Mental Retardation 1 gene |
| FXS | Fragile X syndrome |
| SPR | Surface plasmon resonance |
| RU | Resonance Units |
| PHENIX | Python-based Hierarchical ENvironment for Integrated Xtallography |
References
- Budworth, H.; McMurray, C.T. A brief history of triplet repeat diseases. Methods Mol. Biol. 2013, 1010, 3–17. [Google Scholar] [PubMed]
- Mirkin, S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006, 16, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.Y.; Chang, C.K.; Kao, Y.F.; Chin, C.H.; Ni, C.W.; Hsu, H.Y.; Hu, N.J.; Hsieh, L.C.; Chou, S.H.; Lee, I.R.; et al. Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc. Natl. Acad. Sci. USA 2017, 114, 9535–9540. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Napierala, M.; Wells, R.D. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 2015, 84, 199–226. [Google Scholar] [CrossRef] [PubMed]
- Satange, R.; Chang, C.K.; Hou, M.H. A survey of recent unusual high-resolution DNA structures provoked by mismatches, repeats and ligand binding. Nucleic Acids Res. 2018, 46, 6416–6434. [Google Scholar] [CrossRef] [PubMed]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
- Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Pieretti, M.; Sutcliffe, J.S.; Richards, S.; Verkerk, A.J.; Holden, J.J.; Fenwick, R.G., Jr.; Warren, S.T.; et al. Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell 1991, 67, 1047–1058. [Google Scholar] [CrossRef]
- Chen, Y.W.; Jhan, C.R.; Neidle, S.; Hou, M.H. Structural basis for the identification of an i-motif tetraplex core with a parallel-duplex junction as a structural motif in CCG triplet repeats. Angew. Chem. Int. Ed. 2014, 53, 10682–10686. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, K.; Hagihara, S.; Goto, Y.; Kobori, A.; Hagihara, M.; Hayashi, G.; Kyo, M.; Nomura, M.; Mishima, M.; Kojima, C. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat. Chem. Biol. 2005, 1, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Hashem, V.I.; Pytlos, M.J.; Klysik, E.A.; Tsuji, K.; Khajavi, M.; Ashizawa, T.; Sinden, R.R. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 2004, 32, 6334–6346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, Y.S.; Tseng, W.H.; Chuang, C.Y.; Hou, M.H. The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013, 41, 4284–4294. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.H.; Robinson, H.; Gao, Y.G.; Wang, A.H. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002, 30, 4910–4917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slavik, M.; Carter, S.K. Chromomycin A3, mithramycin, and olivomycin: Antitumor antibiotics of related structure. Adv. Pharmacol. Chemother. 1975, 12, 1–30. [Google Scholar] [PubMed]
- Hsu, C.W.; Chuang, S.M.; Wu, W.L.; Hou, M.H. The crucial role of divalent metal ions in the DNA-acting efficacy and inhibition of the transcription of dimeric chromomycin A3. PLoS ONE 2012, 7, e43792. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.H.; Chang, C.K.; Wu, P.C.; Hu, N.J.; Lee, G.H.; Tzeng, C.C.; Neidle, S.; Hou, M.H. Induced-fit recognition of CCG trinucleotide repeats by a nickel-chromomycin complex resulting in large-scale DNA deformation. Angew. Chem. Int. Ed. 2017, 56, 8761–8765. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA. Discovery Studio Client; 2.5.0.9164; Dassault Systèmes: San Diego, CA, USA, 2005. [Google Scholar]
- Gao, X.; Huang, X.; Smith, G.K.; Zheng, M.; Liu, H. New antiparallel duplex motif of DNA CCG repeats that is stabilized by extrahelical bases symmetrically located in the minor groove. J. Am. Chem. Soc. 1995, 117, 8883–8884. [Google Scholar] [CrossRef]
- Zheng, M.; Huang, X.; Smith, G.K.; Yang, X.; Gao, X. Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J. Mol. Biol. 1996, 264, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Zhang, Y.; Man, V.H.; Roland, C.; Sagui, C. E-motif formed by extrahelical cytosine bases in DNA homoduplexes of trinucleotide and hexanucleotide repeats. Nucleic Acids Res. 2018, 46, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Konig, S.L.; Huppert, J.L.; Sigel, R.K.; Evans, A.C. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013, 41, 7453–7461. [Google Scholar] [CrossRef] [PubMed]
- Fojtik, P.; Vorlickova, M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001, 29, 4684–4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorlickova, M.; Zimulova, M.; Kovanda, J.; Fojtik, P.; Kypr, J. Conformational properties of DNA dodecamers containing four tandem repeats of the CNG triplets. Nucleic Acids Res. 1998, 26, 2679–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, D.; Wilson, W.D.; Neidle, S. Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster. J. Am. Chem. Soc. 2013, 135, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Erlitzki, N.; Huang, K.; Xhani, S.; Farahat, A.A.; Kumar, A.; Boykin, D.W.; Poon, G.M.K. Investigation of the electrostatic and hydration properties of DNA minor groove-binding by a heterocyclic diamidine by osmotic pressure. Biophys. Chem. 2017, 231, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Warren, S.T. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 2000, 9, 901–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigsby, J. The fragile X mental retardation 1 gene (FMR1): Historical perspective, phenotypes, mechanism, pathology, and epidemiology. Clin. Neuropsychol. 2016, 30, 815–833. [Google Scholar] [CrossRef] [PubMed]
- Zamiri, B.; Mirceta, M.; Bomsztyk, K.; Macgregor, R.B., Jr.; Pearson, C.E. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8*(GGCCCC)8 repeat: Effect of CpG methylation. Nucleic Acids Res. 2015, 43, 10055–10064. [Google Scholar] [PubMed]
- Moore, H.; Greenwell, P.W.; Liu, C.P.; Arnheim, N.; Petes, T.D. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 1999, 96, 1504–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fojtik, P.; Kejnovska, I.; Vorlickova, M. The guanine-rich fragile X chromosome repeats are reluctant to form tetraplexes. Nucleic Acids Res. 2004, 32, 298–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirkin, S.M. Expandable DNA repeats and human disease. Nature 2007, 447, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Gan, J.; Huang, Z. Structure-based DNA-targeting strategies with small molecule ligands for drug discovery. Med. Res. Rev. 2013, 33, 1119–1173. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.-H.; Satange, R.; Chang, C.-K. Chapter 6 Binding of small molecules to trinucleotide DNA repeats associated with neurodegenerative diseases. In DNA-targeting Molecules as Therapeutic Agents; Waring, M.J., Ed.; Royal Society of Chemistry: London, UK, 2018; pp. 144–174. [Google Scholar]
- Chang, C.K.; Jhan, C.R.; Hou, M.H. The interaction of DNA-binding ligands with trinucleotide-repeat DNA: Implications for therapy and diagnosis of neurological disorders. Curr. Top. Med. Chem. 2015, 15, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Lu, X.J.; Olson, W.K. Web 3DNA—A web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009, 37, W240–W246. [Google Scholar] [CrossRef] [PubMed]
- Turel, I.; Kljun, J. Interactions of metal ions with DNA, its constituents and derivatives, which may be relevant for anticancer research. Curr. Top. Med. Chem. 2011, 11, 2661–2687. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.G.; Sriram, M.; Wang, A.H. Crystallographic studies of metal ion-DNA interactions: Different binding modes of cobalt(II), copper(II) and barium(II) to N7 of guanines in Z-DNA and a drug-DNA complex. Nucleic Acids Res. 1993, 21, 4093–4101. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.L., Jr. DNA-bound metal ions: Recent developments. Biomol. Concepts 2014, 5, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Theophanides, T.; Anastassopoulou, J. The effects of metal ion contaminants on the double stranded DNA helix and diseases. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2017, 52, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Egli, M. DNA-cation interactions: Quo vadis? Chem. Biol. 2002, 9, 277–286. [Google Scholar] [CrossRef]
- Eichhorn, G.L.; Shin, Y.A. Interaction of metal ions with polynucleotides and related compounds. XII. The relative effect of various metal ions on DNA helicity. J. Am. Chem. Soc. 1968, 90, 7323–7328. [Google Scholar] [CrossRef] [PubMed]
- Gochin, M. A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance. Structure 2000, 8, 441–452. [Google Scholar] [CrossRef]
- Day, H.A.; Wright, E.P.; MacDonald, C.J.; Gates, A.J.; Waller, Z.A. Reversible DNA i-motif to hairpin switching induced by copper(II) cations. Chem. Commun. 2015, 51, 14099–14102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, J.; Yamada, T.; Hirose, C.; Okamoto, I.; Tanaka, Y.; Ono, A. Crystal structure of metallo DNA duplex containing consecutive Watson-Crick-like T-Hg(II)-T base pairs. Angew. Chem. Int. Ed. 2014, 53, 2385–2388. [Google Scholar] [CrossRef] [PubMed]
- Cardin, C.J.; Kelly, J.M.; Quinn, S.J. Photochemically active DNA-intercalating ruthenium and related complexes–insights by combining crystallography and transient spectroscopy. Chem. Sci. 2017, 8, 4705–4723. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Hou, M.H. The binding of the Co(II) complex of dimeric chromomycin A3 to GC sites with flanking G:G mismatches. J. Inorg. Biochem. 2013, 121, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.H.; Robinson, H.; Gao, Y.G.; Wang, A.H. Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion. Nucleic Acids Res. 2004, 32, 2214–2222. [Google Scholar] [CrossRef] [PubMed]
- Cantor, C.R.; Tinoco, I., Jr. Absorption and optical rotatory dispersion of seven trinucleoside diphosphates. J. Mol. Biol. 1965, 13, 65–77. [Google Scholar] [CrossRef]
- Hou, M.H.; Lu, W.J.; Huang, C.Y.; Fan, R.J.; Yuann, J.M. Effects of polyamines on the DNA-reactive properties of dimeric mithramycin complexed with cobalt(II): Implications for anticancer therapy. Biochemistry 2009, 48, 4691–4698. [Google Scholar] [CrossRef] [PubMed]
- Yuann, J.M.; Tseng, W.H.; Lin, H.Y.; Hou, M.H. The effects of loop size on Sac7d-hairpin DNA interactions. Biochim. Biophys. Acta Proteins Proteom. 2012, 1824, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.; Vojtechovsky, J.; Clowney, L.; Brunger, A.T.; Berman, H.M. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 57–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 2 July 2018).
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]





| Drugs | DNA Forms | ka (M−1s−1) | kd (s−1) | Ka (M−1) |
|---|---|---|---|---|
| CoII(Chro)2 | CCG2 | null a | null a | null a |
| CCG3 | 4.19 × 103 | 4.54 × 10−2 | 9.12 × 104 | |
| CCG4 | 8.15 × 103 | 5.68 × 10−2 | 1.43 × 105 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-W.; Satange, R.; Wu, P.-C.; Jhan, C.-R.; Chang, C.-k.; Chung, K.-R.; Waring, M.J.; Lin, S.-W.; Hsieh, L.-C.; Hou, M.-H. CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex. Int. J. Mol. Sci. 2018, 19, 2796. https://doi.org/10.3390/ijms19092796
Chen Y-W, Satange R, Wu P-C, Jhan C-R, Chang C-k, Chung K-R, Waring MJ, Lin S-W, Hsieh L-C, Hou M-H. CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex. International Journal of Molecular Sciences. 2018; 19(9):2796. https://doi.org/10.3390/ijms19092796
Chicago/Turabian StyleChen, Yu-Wen, Roshan Satange, Pei-Ching Wu, Cyong-Ru Jhan, Chung-ke Chang, Kuang-Ren Chung, Michael J. Waring, Sheng-Wei Lin, Li-Ching Hsieh, and Ming-Hon Hou. 2018. "CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex" International Journal of Molecular Sciences 19, no. 9: 2796. https://doi.org/10.3390/ijms19092796
APA StyleChen, Y. -W., Satange, R., Wu, P. -C., Jhan, C. -R., Chang, C. -k., Chung, K. -R., Waring, M. J., Lin, S. -W., Hsieh, L. -C., & Hou, M. -H. (2018). CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex. International Journal of Molecular Sciences, 19(9), 2796. https://doi.org/10.3390/ijms19092796