Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis
Abstract
:1. Introduction
2. Results
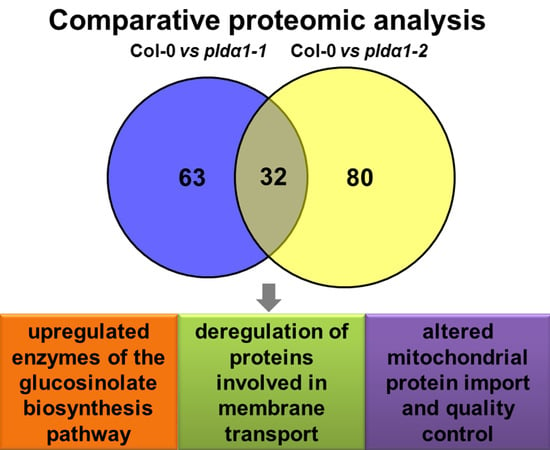
2.1. Overview of Differential Root Proteomes in Two pldα1 Mutants
2.2. Classification of Root Differential Proteomes in pldα1 Mutants
3. Discussion
3.1. New Insights into ABA Signalling
3.2. Mitochondrial Protein Import and Quality Control
3.3. Vesicular Transport
3.4. Glucosinolate Biosynthesis
4. Materials and Methods
4.1. Plant Material
4.2. Protein Extraction and Trypsin Digestion
4.3. Liquid Chromatography, Mass Spectrometry, Protein Identification and Relative Quantitative Analysis
4.4. Bioinformatic Analysis
4.5. Immunoblotting Analysis
4.6. Whole Mount Immunofluorescence Labelling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, X. (Ed.) Phospholipases in Plant Signaling; Signaling and Communication in Plants; Springer: Berlin/Heidelberg, Germany, 2014; Volume 20, ISBN 978-3-642-42010-8. [Google Scholar]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.-C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Lin, F.; Mao, T.; Nie, J.; Yan, M.; Yuan, M.; Zhang, W. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 2012, 24, 4555–4576. [Google Scholar] [CrossRef] [PubMed]
- Testerink, C.; Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 2011, 62, 2349–2361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Qin, C.; Zhao, J.; Wang, X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 9508–9513. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, S.P.; Roth, M.R.; Baughman, E.; Li, M.; Tamura, P.; Jeannotte, R.; Welti, R.; Wang, X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry 2006, 67, 1907–1924. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-B.; Chu, Y.-J.; Xue, H.-W. Phosphatidic Acid (PA) Binds PP2AA1 to Regulate PP2A Activity and PIN1 Polar Localization. Mol. Plant 2013, 6, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Boutté, Y.; Moreau, P. Modulation of endomembranes morphodynamics in the secretory/retrograde pathways depends on lipid diversity. Curr. Opin. Plant Biol. 2014, 22, 22–29. [Google Scholar] [CrossRef]
- Dhonukshe, P.; Laxalt, A.M.; Goedhart, J.; Gadella, T.W.J.; Munnik, T. Phospholipase d activation correlates with microtubule reorganization in living plant cells. Plant Cell 2003, 15, 2666–2679. [Google Scholar] [CrossRef]
- Pleskot, R.; Potocký, M.; Pejchar, P.; Linek, J.; Bezvoda, R.; Martinec, J.; Valentová, O.; Novotná, Z.; Zárský, V. Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 2010, 62, 494–507. [Google Scholar] [CrossRef] [Green Version]
- Pleskot, R.; Li, J.; Žárský, V.; Potocký, M.; Staiger, C.J. Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 2013, 18, 496–504. [Google Scholar] [CrossRef]
- Novák, D.; Vadovič, P.; Ovečka, M.; Šamajová, O.; Komis, G.; Colcombet, J.; Šamaj, J. Gene expression pattern and protein localization of Arabidopsis Phospholipase D Alpha 1 revealed by advanced light-sheet and super-resolution microscopy. Front. Plant Sci. 2018, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.; Zhang, W.; Deng, F.; Zhao, J.; Wang, X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 2006, 312, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Mishra, G.; Markham, J.E.; Li, M.; Tawfall, A.; Welti, R.; Wang, X. Connections between sphingosine kinase and phospholipase D in the abscisic acid signaling pathway in Arabidopsis. J. Biol. Chem. 2012, 287, 8286–8296. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Pandey, S. The role of PLDα1 in providing specificity to signal-response coupling by heterotrimeric G-protein components in Arabidopsis. Plant J. 2016, 86, 50–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy Choudhury, S.; Pandey, S. Phosphatidic acid binding inhibits RGS1 activity to affect specific signaling pathways in Arabidopsis. Plant J. 2017, 90, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zheng, S.; Wang, X. Dual functions of phospholipase Dalpha1 in plant response to drought. Mol. Plant 2008, 1, 262–269. [Google Scholar] [CrossRef]
- Huo, C.; Zhang, B.; Wang, H.; Wang, F.; Liu, M.; Gao, Y.; Zhang, W.; Deng, Z.; Sun, D.; Tang, W. Comparative Study of Early Cold-Regulated Proteins by Two-Dimensional Difference Gel Electrophoresis Reveals a Key Role for Phospholipase Dα1 in Mediating Cold Acclimation Signaling Pathway in Rice. Mol. Cell. Proteom. 2016, 15, 1397–1411. [Google Scholar] [CrossRef]
- Lu, S.; Bahn, S.C.; Qu, G.; Qin, H.; Hong, Y.; Xu, Q.; Zhou, Y.; Hong, Y.; Wang, X. Increased expression of phospholipase Dα1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol. J. 2013, 11, 380–389. [Google Scholar] [CrossRef]
- Uraji, M.; Katagiri, T.; Okuma, E.; Ye, W.; Hossain, M.A.; Masuda, C.; Miura, A.; Nakamura, Y.; Mori, I.C.; Shinozaki, K.; et al. Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2012, 159, 450–460. [Google Scholar] [CrossRef]
- Devaiah, S.P.; Pan, X.; Hong, Y.; Roth, M.; Welti, R.; Wang, X. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis: Phospholipase D in seed aging. Plant J. 2007, 50, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Ufer, G.; Gertzmann, A.; Gasulla, F.; Röhrig, H.; Bartels, D. Identification and characterization of the phosphatidic acid-binding A. thaliana phosphoprotein PLDrp1 that is regulated by PLDα1 in a stress-dependent manner. Plant J. 2017, 92, 276–290. [Google Scholar] [CrossRef]
- Gajiwala, K.S.; Burley, S.K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000, 10, 110–116. [Google Scholar] [CrossRef]
- Rodriguez, L.; Gonzalez-Guzman, M.; Diaz, M.; Rodrigues, A.; Izquierdo-Garcia, A.C.; Peirats-Llobet, M.; Fernandez, M.A.; Antoni, R.; Fernandez, D.; Marquez, J.A.; et al. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. Plant Cell 2014, 26, 4802–4820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Berkey, R.; Blakeslee, J.J.; Lin, J.; Ma, X.; King, H.; Liddle, A.; Guo, L.; Munnik, T.; Wang, X.; et al. Arabidopsis phospholipase Dα1 and Dδ oppositely modulate EDS1- and SA-independent basal resistance against adapted powdery mildew. J. Exp. Bot. 2018, 69, 3675–3688. [Google Scholar] [CrossRef] [PubMed]
- Pleskot, R.; Pejchar, P.; Staiger, C.J.; Potocký, M. When fat is not bad: The regulation of actin dynamics by phospholipid signaling molecules. Front. Plant Sci. 2014, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Steinborn, K. The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 2002, 16, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Cui, M.; Qian, D.; Zhu, L.; Wei, C.; Yuan, M.; Zhang, Z.; Li, Y. AtTFC B is involved in control of cell division. Front. Biosci. Elite Ed. 2010, 2, 752–763. [Google Scholar]
- Dhonukshe, P.; Bargmann, B.O.R.; Gadella, T.W.J. Arabidopsis Tubulin Folding Cofactor B Interacts with α-Tubulin In Vivo. Plant Cell Physiol. 2006, 47, 1406–1411. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Deng, Z.; Paredez, A.R.; DeBolt, S.; Wang, Z.-Y.; Somerville, C. Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 18064–18069. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, N.; Sarkeshik, A.; Nito, K.; Park, S.-Y.; Wang, A.; Carvalho, P.C.; Lee, S.; Caddell, D.F.; Cutler, S.R.; Chory, J.; et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. Cell Mol. Biol. 2010, 61, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Postaire, O.; Tournaire-Roux, C.; Grondin, A.; Boursiac, Y.; Morillon, R.; Schäffner, A.R.; Maurel, C. A PIP1 aquaporin contributes to hydrostatic pressure-induced water transport in both the root and rosette of Arabidopsis. Plant Physiol. 2010, 152, 1418–1430. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Arisz, S.A.; Dekker, H.L.; Kramer, G.; de Koster, C.G.; Haring, M.A.; Munnik, T.; Testerink, C. Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 2013, 450, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Bellati, J.; Champeyroux, C.; Hem, S.; Rofidal, V.; Krouk, G.; Maurel, C.; Santoni, V. Novel Aquaporin Regulatory Mechanisms Revealed by Interactomics. Mol. Cell. Proteom. 2016, 15, 3473–3487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Carrie, C.; Giraud, E.; Elhafez, D.; Narsai, R.; Duncan, O.; Whelan, J.; Murcha, M.W. Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 2012, 24, 2675–2695. [Google Scholar] [CrossRef] [PubMed]
- Neupert, W. A Perspective on Transport of Proteins into Mitochondria: A Myriad of Open Questions. J. Mol. Biol. 2015, 427, 1135–1158. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.F.; Glaser, E. Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 360–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Aken, O.; Whelan, J.; Van Breusegem, F. Prohibitins: Mitochondrial partners in development and stress response. Trends Plant Sci. 2010, 15, 275–282. [Google Scholar] [CrossRef]
- Piechota, J.; Bereza, M.; Sokołowska, A.; Suszyński, K.; Lech, K.; Jańska, H. Unraveling the functions of type II-prohibitins in Arabidopsis mitochondria. Plant Mol. Biol. 2015, 88, 249–267. [Google Scholar] [CrossRef]
- Ahn, C.S.; Lee, J.H.; Reum Hwang, A.; Kim, W.T.; Pai, H.-S. Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 2006, 46, 658–667. [Google Scholar] [CrossRef] [Green Version]
- Donaldson, J.G. Phospholipase D in endocytosis and endosomal recycling pathways. Biochim. Biophys. Acta 2009, 1791, 845–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonescu, C.N.; Danuser, G.; Schmid, S.L. Phosphatidic Acid Plays a Regulatory Role in Clathrin-mediated Endocytosis. Mol. Biol. Cell 2010, 21, 2944–2952. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Xue, H.-W. Arabidopsis PLD 2 Regulates Vesicle Trafficking and Is Required for Auxin Response. Plant Cell 2007, 19, 281–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, M.G. Molecular mechanisms of PLD function in membrane traffic. Traffic 2008, 9, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.; Panda, A.; Coessens, E.; Raj, N.; Yadav, S.; Balakrishnan, S.; Zhang, Q.; Georgiev, P.; Basak, B.; Pasricha, R.; et al. Phospholipase D activity couples plasma membrane endocytosis with retromer dependent recycling. eLife 2016, 5, e18515. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, K.; Krebs, M. The V-ATPase: Small cargo, large effects. Curr. Opin. Plant Biol. 2010, 13, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Uemura, T.; Ebine, K.; Nishimori, Y.; Ueda, T.; Nakano, A.; Sato, M.H.; Fukao, Y. Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.; Wickner, W.; Haas, A. Sec18p (NSF)-Driven Release of Sec17p (α-SNAP) Can Precede Docking and Fusion of Yeast Vacuoles. Cell 1996, 85, 83–94. [Google Scholar] [CrossRef]
- Wang, T.; Li, L.; Hong, W. SNARE proteins in membrane trafficking. Traffic 2017, 18, 767–775. [Google Scholar] [CrossRef]
- Uemura, T.; Ueda, T.; Ohniwa, R.L.; Nakano, A.; Takeyasu, K.; Sato, M.H. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 2004, 29, 49–65. [Google Scholar] [CrossRef]
- Wang, C.; Yan, X.; Chen, Q.; Jiang, N.; Fu, W.; Ma, B.; Liu, J.; Li, C.; Bednarek, S.Y.; Pan, J. Clathrin Light Chains Regulate Clathrin-Mediated Trafficking, Auxin Signaling, and Development in Arabidopsis. Plant Cell 2013, 25, 499–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, L.; Li, R.; Pan, J.; Ding, Z.; Lin, J. Endocytosis and its regulation in plants. Trends Plant Sci. 2015, 20, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, S.; Shimono, Y.; Shirakawa, M.; Fukao, Y.; Kawase, T.; Hatsugai, N.; Tamura, K.; Shimada, T.; Hara-Nishimura, I. Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 2013, 25, 2958–2969. [Google Scholar] [CrossRef] [PubMed]
- Zelazny, E.; Santambrogio, M.; Pourcher, M.; Chambrier, P.; Berne-Dedieu, A.; Fobis-Loisy, I.; Miège, C.; Jaillais, Y.; Gaude, T. Mechanisms Governing the Endosomal Membrane Recruitment of the Core Retromer in Arabidopsis. J. Biol. Chem. 2013, 288, 8815–8825. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, S.Y.; Song, K.; Sohn, E.J.; Lee, Y.; Lee, D.W.; Hara-Nishimura, I.; Hwang, I. Trafficking of Vacuolar Proteins: The Crucial Role of Arabidopsis Vacuolar Protein Sorting 29 in Recycling Vacuolar Sorting Receptor. Plant Cell 2012, 24, 5058–5073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nodzyński, T.; Feraru, M.I.; Hirsch, S.; De Rycke, R.; Niculaes, C.; Boerjan, W.; Van Leene, J.; De Jaeger, G.; Vanneste, S.; Friml, J. Retromer Subunits VPS35A and VPS29 Mediate Prevacuolar Compartment (PVC) Function in Arabidopsis. Mol. Plant 2013, 6, 1849–1862. [Google Scholar] [CrossRef]
- Hino, T.; Tanaka, Y.; Kawamukai, M.; Nishimura, K.; Mano, S.; Nakagawa, T. Two Sec13p Homologs, AtSec13A and AtSec13B, Redundantly Contribute to the Formation of COPII Transport Vesicles in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2011, 75, 1848–1852. [Google Scholar] [CrossRef]
- Fridmann-Sirkis, Y.; Siniossoglou, S.; Pelham, H.R.B. TMF is a golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 2004, 5, 18. [Google Scholar] [CrossRef]
- Latijnhouwers, M.; Gillespie, T.; Boevink, P.; Kriechbaumer, V.; Hawes, C.; Carvalho, C.M. Localization and domain characterization of Arabidopsis golgin candidates. J. Exp. Bot. 2007, 58, 4373–4386. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Richardson, C.; Hawkins, T.J.; Sparkes, I.; Hawes, C.; Hussey, P.J. Plant VAP27 proteins: Domain characterization, intracellular localization and role in plant development. New Phytol. 2016, 210, 1311–1326. [Google Scholar] [CrossRef]
- Saravanan, R.S.; Slabaugh, E.; Singh, V.R.; Lapidus, L.J.; Haas, T.; Brandizzi, F. The targeting of the oxysterol-binding protein ORP3a to the endoplasmic reticulum relies on the plant VAP33 homolog PVA12. Plant J. Cell Mol. Biol. 2009, 58, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Siao, W.; Wang, P.; Voigt, B.; Hussey, P.J.; Baluska, F. Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum–plasma membrane contact sites. J. Exp. Bot. 2016, 67, 6161–6171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takáč, T.; Pechan, T.; Richter, H.; Müller, J.; Eck, C.; Böhm, N.; Obert, B.; Ren, H.; Niehaus, K.; Šamaj, J. Proteomics on brefeldin A-treated Arabidopsis roots reveals profilin 2 as a new protein involved in the cross-talk between vesicular trafficking and the actin cytoskeleton. J. Proteome Res. 2011, 10, 488–501. [Google Scholar] [CrossRef]
- Takáč, T.; Pechan, T.; Šamajová, O.; Ovečka, M.; Richter, H.; Eck, C.; Niehaus, K.; Šamaj, J. Wortmannin treatment induces changes in Arabidopsis root proteome and post-Golgi compartments. J. Proteome Res. 2012, 11, 3127–3142. [Google Scholar] [CrossRef] [PubMed]
- Janda, M.; Šašek, V.; Chmelařová, H.; Andrejch, J.; Nováková, M.; Hajšlová, J.; Burketová, L.; Valentová, O. Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 59. [Google Scholar] [CrossRef]
- Kravets, V.S.; Kretinin, S.V.; Kolesnikov, Y.S.; Getman, I.A.; Romanov, G.A. Cytokinins evoke rapid activation of phospholipase D in sensitive plant tissues. Dokl. Biochem. Biophys. 2009, 428, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zheng, S.; Wang, X. Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 1997, 9, 2183. [Google Scholar] [CrossRef]
- Testerink, C.; Larsen, P.B.; van der Does, D.; van Himbergen, J.A.J.; Munnik, T. Phosphatidic acid binds to and inhibits the activity of Arabidopsis CTR1. J. Exp. Bot. 2007, 58, 3905–3914. [Google Scholar] [CrossRef] [Green Version]
- Malka, S.K.; Cheng, Y. Possible Interactions between the Biosynthetic Pathways of Indole Glucosinolate and Auxin. Front. Plant Sci. 2017, 8, 2131. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J.; et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 47, 10–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, A.; Wang, C.; Crocoll, C.; Halkier, B.A. Biotechnological approaches in glucosinolate production. J. Integr. Plant Biol. 2018, 60, 1231–1248. [Google Scholar] [CrossRef] [PubMed]
- Takáč, T.; Šamajová, O.; Pechan, T.; Luptovčiak, I.; Šamaj, J. Feedback microtubule control and microtubule-actin cross-talk in arabidopsis revealed by integrative proteomic and cell biology analysis of KATANIN 1 mutants. Mol. Cell. Proteom. 2017, 16, 1591–1609. [Google Scholar] [CrossRef] [PubMed]
- Takáč, T.; Vadovič, P.; Pechan, T.; Luptovčiak, I.; Šamajová, O.; Šamaj, J. Comparative proteomic study of Arabidopsis mutants mpk4 and mpk6. Sci. Rep. 2016, 6, 28306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Tsuji, J.; Fu, S.-C.; Tomii, K.; Horton, P.; Imai, K. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Šamajová, O.; Komis, G.; Šamaj, J. Immunofluorescent Localization of MAPKs and Colocalization with Microtubules in Arabidopsis Seedling Whole-Mount Probes. In Plant MAP Kinases: Methods and Protocols; Komis, G., Šamaj, J., Eds.; Springer: New York, NY, USA, 2014; pp. 107–115. ISBN 978-1-4939-0922-3. [Google Scholar]







| TAIR Accession Number | UNIPROT Accession Number | Sequence Name | pldα1-1/Col-0 Ratio | pldα1-2/Col-0 Ratio | pldα1-1/Col-0 p Value | pldα1-2/Col-0 p Value |
|---|---|---|---|---|---|---|
| Translation | ||||||
| Q8LD46 | At2g39460 | 60S ribosomal protein L23a-1 | 20.82 | 7.62 | 0.01 | 0.012 |
| Q9LHG9 | At3g12390 | Nascent polypeptide-associated complex subunit alpha-like protein 1 | 1.82 | 1.91 | 0.052 | 0.029 |
| Q9FJH6 | At5g60790 | ABC transporter F family member 1 | Unique in WT | 0.38 | n.a. | 0.03 |
| Stress response | ||||||
| P50700 | At4g11650 | Osmotin-like protein OSM34 | 0.42 | 0.26 | 0.048 | 0.039 |
| Q9LYW9 | At5g03160 | DnaJ protein P58IPK homolog | 4.03 | 4.11 | 0.004 | 0.026 |
| P24102 | At2g38380 | Peroxidase 22 | 1.79 | 1.99 | 0.031 | 0.005 |
| Q9LSY7 | At3g21770 | Peroxidase 30 | Unique in mutant | Unique in mutant | n.a. | n.a. |
| P42760 | At1g02930 | Glutathione S-transferase F6 | 0.29 | 0.26 | 0.049 | 0.029 |
| Q9SRY5 | At1g02920 | Glutathione S-transferase F7 | 0.31 | 0.25 | 0.053 | 0.036 |
| Q38882 | At3g15730 | Phospholipase D alpha 1 | Unique in WT | Unique in WT | n.a. | n.a. |
| Q9FKA5 | At5g39570 | Uncharacterized protein At5g39570 (PLD regulated protein1, PLDRP1) | Unique in WT | Unique in WT | n.a. | n.a. |
| P32961 | At3g44310 | Nitrilase 1 | 1.79 | 1.79 | 0.018 | 0.023 |
| Membrane transport | ||||||
| Q9SRI1 | At3g01340 | Protein transport protein SEC13 homolog A | 2.55 | 2.97 | 0.001 | 0.011 |
| Q8S9J8 | At4g32285 | Probable clathrin assembly protein At4g32285 | Unique in WT | Unique in WT | n.a. | n.a. |
| Mitochondrial respiratory chain | ||||||
| Q9FT52 | At3g52300 | ATP synthase subunit d, mitochondrial | 1.69 | 1.57 | 0.047 | 0.046 |
| O81845 | At3g54110 | Mitochondrial uncoupling protein 1 | 1.67 | 2.14 | 0.02 | 0.01 |
| P93306 | AtMg00510 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 | 0.50 | 0.56 | 0.028 | 0.054 |
| Q9S7L9 | At1g22450 | Cytochrome c oxidase subunit 6b-1 | Unique in mutant | Unique in mutant | n.a. | n.a. |
| Glucosinolate biosynthesis | ||||||
| O49340 | At2g30750 | Cytochrome P450 71A12 | Unique in WT | Unique in WT | n.a. | n.a. |
| Q9FG67 | At5g23010 | Methylthioalkylmalate synthase 1, chloroplastic | 1.13 | 1.74 | 0.01 | 0.036 |
| Other functions | ||||||
| Q9LSB4 | At3g15950 | TSA1-like protein | 1.33 | 1.71 | 0.049 | 0.003 |
| Q9SP02 | At5g58710 | Peptidyl-prolyl cis-trans isomerase CYP20-1 | 1.13 | 1.56 | 0.004 | 0.013 |
| Q8VYV7 | At5g66120 | 3-dehydroquinate synthase, chloroplastic | 0.39 | 0.44 | 0.046 | 0.01 |
| Q9AV97 | At1g79500 | 2-dehydro-3-deoxyphosphooctonate aldolase 1 | Unique in mutant | Unique in mutant | n.a. | n.a. |
| Q9FHR8 | At5g43280 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, peroxisomal | 0.44 | 0.30 | 0.011 | 0.003 |
| Q9FIK7 | At5g47720 | Probable acetyl-CoA acetyltransferase, cytosolic 2 | 0.59 | 1.71 | 0.042 | 0.055 |
| Q9FLQ4 | At5g55070 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 1, mitochondrial | 8.28 | 5.54 | 0.008 | 0.029 |
| Q9FMT1 | At5g14200 | 3-isopropylmalate dehydrogenase 3, chloroplastic | 1.49 | 1.61 | 0.002 | 0.01 |
| Q9LQ04 | At1g63000 | Bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase | 1.38 | 1.60 | 0.038 | 0.029 |
| Q9SA14 | At1g31180 | 3-isopropylmalate dehydrogenase 1, chloroplastic | 1.52 | 1.55 | 0.019 | 0.02 |
| Q9SIU0 | At2g13560 | NAD-dependent malic enzyme 1, mitochondrial | 4.99 | 2.10 | 0.011 | 0.009 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takáč, T.; Šamajová, O.; Vadovič, P.; Pechan, T.; Šamaj, J. Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis. Int. J. Mol. Sci. 2019, 20, 82. https://doi.org/10.3390/ijms20010082
Takáč T, Šamajová O, Vadovič P, Pechan T, Šamaj J. Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis. International Journal of Molecular Sciences. 2019; 20(1):82. https://doi.org/10.3390/ijms20010082
Chicago/Turabian StyleTakáč, Tomáš, Olga Šamajová, Pavol Vadovič, Tibor Pechan, and Jozef Šamaj. 2019. "Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis" International Journal of Molecular Sciences 20, no. 1: 82. https://doi.org/10.3390/ijms20010082
APA StyleTakáč, T., Šamajová, O., Vadovič, P., Pechan, T., & Šamaj, J. (2019). Shot-Gun Proteomic Analysis on Roots of Arabidopsis pldα1 Mutants Suggesting the Involvement of PLDα1 in Mitochondrial Protein Import, Vesicular Trafficking and Glucosinolate Biosynthesis. International Journal of Molecular Sciences, 20(1), 82. https://doi.org/10.3390/ijms20010082