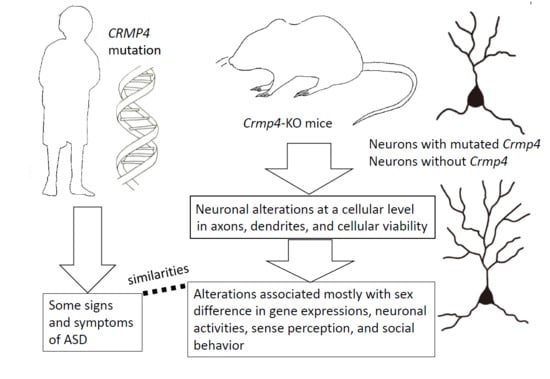
Crmp4-KO Mice as an Animal Model for Investigating Certain Phenotypes of Autism Spectrum Disorders
Abstract
:1. Introduction
2. Identification of CRMP4
3. The Regulatory Mechanisms Suggested for CRMP4
4. Potential Involvement of CRMPs Including CRMP4 in Neurodevelopmental Disorders
5. Behavioral and Perceptual Abnormalities Observed in Crmp4-KO Mice
5.1. Impairments in Social Behavior
5.2. Abnormalities in Sensory Perception
6. Altered Dendritic Arborization in Crmp4-KO Mice and Crmp4-Knockdown (KD) Neurons
7. Altered Expressions of Genes Related to Excitatory and Inhibitory Synaptic Transmission in the Brain of Crmp4-KO Mice
8. Sex-Specific Phenotypes Observed in Crmp4-KO Mice and Other Animal Models of ASD
9. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADHD | attention-deficit/hyperactivity disorder |
| AMPA | α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate |
| ASD | autism spectrum disorder |
| AVPV | anteroventral periventricular nucleus |
| CRMP | collapsin response mediator protein |
| DRP | dihydropyrimidase; DPYSL3, dihydropyrimidase-like 3 |
| EA | ethyl acetate |
| EPL | external plexiform layer |
| FMR1 | fragile X mental retardation 1 gene |
| GCL | granule cell layer |
| GL | glomerular layer |
| GluR1 | glutamate receptor 1 |
| GluT1 | glutamate transporter 1 |
| KO | knockout |
| MAI | myelin-associated inhibitor |
| MCL | mitral cell layer |
| NMDA | N-methyl-D-aspartate |
| OB | olfactory bulb |
| RT | room temperature |
| TERT-tg mice | telomerase reverse transcriptase-overexpressing transgenic mice |
| TOAD-64 | turned on after division 64 |
| TUC-4 | TOAD-64/Ulip-1/CRMP4 |
| Ulip-1 | Ulip-1 |
| UV | ultrasonic vocalization |
| vGluT1 | vesicular glutamate transporter 1 |
| WT | wild type |
References
- Charrier, E.; Reibel, S.; Rogemond, V.; Aguera, M.; Thomasset, N.; Honnorat, J. Collapsin response mediator proteins (CRMPs): Involvement in nervous system development and adult neurodegenerative disorders. Mol. Neurol. 2003, 28, 51–64. [Google Scholar] [CrossRef]
- Schmidt, E.F.; Strittmatter, S.M. The CRMP family of proteins and their role in Sema3A signaling. Adv. Exp. Med. Biol. 2007, 600, 1–11. [Google Scholar] [PubMed]
- Quach, T.T.; Honnorat, J.; Kolattukudy, P.E.; Khanna, R.; Duchemin, A.M. CRMPs: Critical molecules for neurite morphogenesis and neuropsychiatric diseases. Mol. Psychiatry. 2015, 20, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Hensley, K.; Venkova, K.; Christov, A.; Gunning, W.; Park, J. Collapsin response mediator protein-2: An emerging pathologic feature and therapeutic target for neurodisease indications. Mol. Neurobiol. 2011, 43, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, N.; Takahashi, A.; Takao, K.; Yamamoto, T.; Kolattukudy, P.; Miyakawa, T.; Goshima, Y. Mice lacking collapsin response mediator protein 1 manifest hyperactivity, impaired learning and memory, and impaired prepulse inhibition. Front. Behav. Neurosci. 2013, 7, 216. [Google Scholar] [CrossRef]
- Lee, H.; Joo, J.; Nah, S.S.; Kim, J.W.; Kim, H.K.; Kwon, J.T.; Lee, H.Y.; Kim, Y.O.; Kim, H.J. Changes in Dpysl2 expression are associated with prenatally stressed rat offspring and susceptibility to schizophrenia in humans. Int. J. Mol. Med. 2015, 35, 1574–1586. [Google Scholar] [CrossRef] [Green Version]
- Pham, X.; Song, G.; Lao, S.; Goff, L.; Zhu, H.; Valle, D.; Avramopoulos, D. The DPYSL2 gene connects mTOR and schizophrenia. Transl. Psychiatry 2016, 6, e933. [Google Scholar] [CrossRef]
- Tsutiya, A.; Nakano, Y.; Hansen-Kiss, E.; Kelly, B.; Nishihara, M.; Goshima, Y.; Corsmeier, D.; White, P.; Herman, G.E.; Ohtani-Kaneko, R. Human CRMP4 mutation and disrupted Crmp4 expression in mice are associated with ASD characteristics and sexual dimorphism. Sci. Rep. 2017, 7, 16812. [Google Scholar] [CrossRef] [Green Version]
- Uchida, Y.; Ohshima, T.; Sasaki, Y.; Suzuki, H.; Yanai, S.; Yamashita, N.; Nakamura, F.; Takei, K.; Ihara, Y.; Mikoshiba, K.; et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: Implication of common phosphorylating mechanism underlying axon guidance and Alzheimer’s disease. Genes Cells 2005, 10, 165–179. [Google Scholar] [CrossRef]
- Toba, J.; Nikkuni, M.; Ishizeki, M.; Yoshii, A.; Watamura, N.; Inoue, T.; Ohshima, T. PPARγ agonist pioglitazone improves cerebellar dysfunction at pre-Aβ deposition stage in APPswe/PS1dE9 Alzheimer’s disease model mice. Biochem. Biophys. Res. Commun. 2016, 473, 1039–1044. [Google Scholar] [CrossRef]
- Kim, A.E.; Kang, P.; Bucelli, R.C.; Ferguson, C.J.; Schmidt, R.E.; Varadhachary, A.S.; Day, G.S. Autoimmune encephalitis with multiple autoantibodies: A diagnostic and therapeutic challenge. Neurologist 2018, 23, 55–59. [Google Scholar] [CrossRef]
- Fujisawa, H.; Ohtani-Kaneko, R.; Naiki, M.; Okada, T.; Masuko, K.; Yudoh, K.; Suematsu, N.; Okamoto, K.; Nishioka, K.; Kato, T. Involvement of post-translational modification of neuronal plasticity-related proteins in hyperalgesia revealed by a proteomic analysis. Proteomics 2008, 8, 1706–1719. [Google Scholar] [CrossRef]
- Piekarz, A.D.; Due, M.R.; Khanna, M.; Wang, B.; Ripsch, M.S.; Wang, R.; Meroueh, S.O.; Vasko, M.R.; White, F.A.; Khanna, R. CRMP-2 peptide mediated decrease of high and low voltage-activated calcium channels, attenuation of nociceptor excitability, and anti-nociception in a model of AIDS therapy-induced painful peripheral neuropathy. Mol. Pain 2012, 8, 54. [Google Scholar] [CrossRef]
- Harada, S.; Matsuura, W.; Takano, M.; Tokuyama, S. Proteomic profiling in the spinal cord and sciatic nerve in a global cerebral ischemia-induced mechanical allodynia mouse model. Biol. Pharm. Bull. 2016, 39, 230–238. [Google Scholar] [CrossRef]
- Lawal, M.F.; Olotu, F.A.; Agoni, C.; Soliman, M.E. Exploring the C-Terminal Tail Dynamics: Structural and Molecular Perspectives into the Therapeutic Activities of Novel CRMP-2 Inhibitors, Naringenin and Naringenin-7-O-glucuronide, in the Treatment of Alzheimer’s Disease. Chem. Biodivers. 2018, 15, e1800437. [Google Scholar] [CrossRef]
- Kolodkin, A.L.; Matthes, D.J.; O’Connor, T.P.; Patel, N.H.; Admon, A.; Bentley, D.; Goodman, C.S. Fasciclin IV: Sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron 1992, 9, 831–845. [Google Scholar] [CrossRef]
- Luo, Y.; Raible, D.; Raper, J.A. Collapsin: A protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 1993, 75, 217–227. [Google Scholar] [CrossRef]
- Raper, J.A. Semaphorins and their receptors in vertebrates and invertebrates. Curr. Opin. Neurobiol. 2000, 10, 88–94. [Google Scholar] [CrossRef]
- Fenstermaker, V.; Chen, Y.; Ghosh, A.; Yuste, R. Regulation of dendritic length and branching by semaphorin 3A. J. Neurobiol. 2004, 58, 403–412. [Google Scholar] [CrossRef] [Green Version]
- Pascual, M.; Pozas, E.; Soriano, E. Role of class 3 semaphorins in the development and maturation of the septohippocampal pathway. Hippocampus 2005, 15, 184–202. [Google Scholar] [CrossRef]
- Yoshida, Y. Semaphorin signaling in vertebrate neural circuit assembly. Front. Mol. Neurosci. 2012, 5, 71. [Google Scholar] [CrossRef] [PubMed]
- Goshima, Y.; Nakamura, F.; Strittmatter, P.; Strittmatter, S.M. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature 1995, 376, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Minturn, J.E.; Fryer, H.J.; Geschwind, D.H.; Hockfield, S. TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J. Neurosci. 1995, 15, 6757–6766. [Google Scholar] [CrossRef]
- Minturn, J.E.; Geschwind, D.H.; Fryer, H.J.; Hockfield, S. Early postmitotic neurons transiently express TOAD-64, a neural specific protein. J. Comp. Neurol. 1995, 355, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Byk, T.; Dobransky, T.; Cifuentes-Diaz, C.; Sobel, A. Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J. Neurosci. 1996, 16, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Hamajima, N.; Matsuda, K.; Sakata, S.; Tamaki, N.; Sasaki, M.; Nonaka, M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene 1996, 180, 157–163. [Google Scholar] [CrossRef]
- Yamashita, N.; Uchida, Y.; Ohshima, T.; Hirai, S.; Nakamura, F.; Taniguchi, M.; Mikoshiba, K.; Honnorat, J.; Kolattukudy, P.; Thomasset, N.; et al. Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J. Neurosci. 2006, 26, 13357–13362. [Google Scholar] [CrossRef]
- Alabed, Y.Z.; Poolm, M.; Tone, S.O.; Sutherland, C.; Fournier, A.E. GSK3 beta regulates myelin-dependent axon outgrowth inhibition through CRMP4. J. Neurosci. 2010, 30, 5635–5643. [Google Scholar] [CrossRef] [PubMed]
- Charrier, E.; Mosinger, B.; Meissirel, C.; Aguera, M.; Rogemond, V.; Reibel, S.; Salin, P.; Chounlamountri, N.; Perrot, V.; Belin, M.F.; et al. Transient alterations in granule cell proliferation, apoptosis and migration in postnatal developing cerebellum of CRMP1−/− mice. Genes Cells 2006, 11, 1337–1352. [Google Scholar] [CrossRef]
- Yamashita, N.; Morita, A.; Uchida, Y.; Nakamura, F.; Usui, H.; Ohshima, T.; Taniguchi, M.; Honnorat, J.; Thomasset, N.; Takei, K.; et al. Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J. Neurosci. 2007, 27, 12546–12554. [Google Scholar] [CrossRef]
- Su, K.Y.; Chien, W.L.; Fu, W.M.; Yu, I.S.; Huang, H.P.; Huang, P.H.; Lin, S.R.; Shih, J.Y.; Lin, Y.L.; Hsueh, Y.P.; et al. Mice deficient in collapsin response mediator protein-1 exhibit impaired long-term potentiation and impaired spatial learning and memory. J. Neurosci. 2007, 27, 2513–2524. [Google Scholar] [CrossRef]
- Yamashita, N.; Goshima, Y. Collapsin response mediator proteins regulate neuronal development and plasticity by switching their phosphorylation status. Mol. Neurobiol. 2012, 45, 234–246. [Google Scholar] [CrossRef]
- Arimura, N.; Inagaki, N.; Chihara, K.; Ménager, C.; Nakamura, N.; Amano, M.; Iwamatsu, A.; Goshima, Y.; Kaibuchi, K. Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J. Biol. Chem. 2000, 275, 23973–23980. [Google Scholar] [CrossRef]
- Arimura, N.; Ménager, C.; Kawano, Y.; Yoshimura, T.; Kawabata, S.; Hattori, A.; Fukata, Y.; Amano, M.; Goshima, Y.; Inagaki, M.; et al. Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol. Cell Biol. 2005, 25, 9973–9984. [Google Scholar] [CrossRef]
- Yoshimura, T.; Kawano, Y.; Arimura, N.; Kawabata, S.; Kikuchi, A.; Kaibuchi, K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 2005, 120, 137–149. [Google Scholar] [CrossRef]
- Cole, A.R.; Causeret, F.; Yadirgi, G.; Hastie, C.J.; McLauchlan, H.; McManus, E.J.; Hernández, F.; Eickholt, B.J.; Nikolic, M.; Sutherland, C. Distinct priming kinases contribute to differential regulation of collapsin response mediator proteins by glycogen synthase kinase-3 in vivo. J. Biol. Chem. 2006, 281, 16591–16598. [Google Scholar] [CrossRef]
- Alabed, Y.Z.; Pool, M.; Ong Tone, S.; Fournier, A.E. Identification of CRMP4 as a convergent regulator of axon outgrowth inhibition. J. Neurosci. 2007, 27, 1702–1711. [Google Scholar] [CrossRef]
- Tanaka, H.; Morimura, R.; Ohshima, T. Dpysl2 (CRMP2) and Dpysl3 (CRMP4) phosphorylation by Cdk5 and DYRK2 is required for proper positioning of Rohon-Beard neurons and neural crest cells during neurulation in zebrafish. Dev. Biol. 2012, 370, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Morimura, R.; Nozawa, K.; Tanaka, H.; Ohshima, T. Phosphorylation of Dpsyl2 (CRMP2) and Dpsyl3 (CRMP4) is required for positioning of caudal primary motor neurons in the zebrafish spinal cord. Dev. Neurobiol. 2013, 73, 911–920. [Google Scholar] [CrossRef] [Green Version]
- Kowara, R.; Chen, Q.; Milliken, M.; Chakravarthy, B. Calpain-mediated truncation of dihydropyrimidinase-like 3 protein (DPYSL3) in response to NMDA and H2O2 toxicity. J. Neurochem. 2005, 95, 466–474. [Google Scholar] [CrossRef]
- Kowara, R.; Moraleja, K.L.; Chakravarthy, B. Involvement of nitric oxide synthase and ROS-mediated activation of L-type voltage-gated Ca2+ channels in NMDA-induced DPYSL3 degradation. Brain Res. 2006, 1119, 40–49. [Google Scholar] [CrossRef]
- Kowara, R.; Moraleja, K.L.; Chakravarthy, B. PLA(2) signaling is involved in calpain-mediated degradation of synaptic dihydropyrimidinase-like 3 protein in response to NMDA excitotoxicity. Neurosci. Lett. 2008, 430, 197–202. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, X.W.; Liu, S.; Hu, K.; Wang, C.; He, Q.; Li, M. Calpain-truncated CRMP-3 and -4 contribute to potassium deprivation-induced apoptosis of cerebellar granule neurons. Proteomics 2009, 9, 3712–3728. [Google Scholar] [CrossRef]
- Quinn, C.C.; Chen, E.; Kinjo, T.G.; Kelly, G.; Bell, A.W.; Elliott, R.C.; McPherson, P.S.; Hockfield, S. TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J. Neurosci. 2003, 23, 2815–2823. [Google Scholar] [CrossRef]
- Yuasa-Kawada, J.; Suzuki, R.; Kano, F.; Ohkawara, T.; Murata, M.; Noda, M. Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur. J. Neurosci. 2003, 17, 2329–2343. [Google Scholar] [CrossRef]
- Tan, M.; Cha, C.; Ye, Y.; Zhang, J.; Li, S.; Wu, F.; Gong, S.; Guo, G. CRMP4 and CRMP2 interact to coordinate cytoskeleton dynamics, regulating growth cone development and axon elongation. Neural. Plast. 2015, 947423. [Google Scholar] [CrossRef]
- Seki, T. Expression patterns of immature neuronal markers PSA-NCAM, CRMP-4 and NeuroD in the hippocampus of young adult and aged rodents. J. Neurosci. Res. 2002, 70, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Cnops, L.; Hu, T.T.; Burnat, K.; Van der Gucht, E.; Arckens, L. Age-dependent alterations in CRMP2 and CRMP4 protein expression profiles in cat visual cortex. Brain Res. 2006, 1088, 109–119. [Google Scholar] [CrossRef]
- Tsutiya, A.; Ohtani-Kaneko, R. Postnatal alteration of collapsin response mediator protein 4 mRNA expression in the mouse brain. J Anat. 2012, 221, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Koide, T.; Aleksic, B.; Ito, Y.; Usui, H.; Yoshimi, A.; Inada, T.; Suzuki, M.; Hashimoto, R.; Takeda, M.; Iwata, N.; et al. A two-stage case-control association study of the dihydropyrimidinase-like 2 gene (DPYSL2) with schizophrenia in Japanese subjects. J. Hum. Genet. 2010, 55, 469–472. [Google Scholar] [CrossRef]
- Bader, V.; Tomppo, L.; Trossbach, S.V.; Bradshaw, N.J.; Prikulis, I.; Leliveld, S.R.; Lin, C.Y.; Ishizuka, K.; Sawa, A.; Ramos, A.; et al. Proteomic, genomic and translational approaches identify CRMP1 for a role in schizophrenia and its underlying traits. Hum. Mol. Genet. 2012, 21, 4406–4418. [Google Scholar] [CrossRef] [Green Version]
- Martins-de-Souza, D.; Cassoli, J.S.; Nascimento, J.M.; Hensley, K.; Guest, P.C.; Pinzon-Velasco, A.M.; Turck, C.W. The protein interactome of collapsin response mediator protein-2 (CRMP2/DPYSL2) reveals novel partner proteins in brain tissue. Proteomics Clin. Appl. 2015, 9, 817–831. [Google Scholar] [CrossRef]
- Liu, Y.; Pham, X.; Zhang, L.; Chen, P.L.; Burzynski, G.; McGaughey, D.M.; He, S.; McGrath, J.A.; Wolyniec, P.; Fallin, M.D.; et al. Functional variants in DPYSL2 sequence increase risk of schizophrenia and suggest a link to mTOR signaling. G3 2014, 5, 61–72. [Google Scholar] [CrossRef]
- Nakamura, H.; Yamashita, N.; Kimura, A.; Kimura, Y.; Hirano, H.; Makihara, H.; Kawamoto, Y.; Jitsuki-Takahashi, A.; Yonezaki, K.; Takase, K.; et al. Comprehensive behavioral study and proteomic analyses of CRMP2-deficient mice. Genes Cells 2016, 21, 1059–1079. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H.; Takahashi-Jitsuki, A.; Makihara, H.; Asano, T.; Kimura, Y.; Nakabayashi, J.; Yamashita, N.; Kawamoto, Y.; Nakamura, F.; Ohshima, T.; et al. Proteome and behavioral alterations in phosphorylation-deficient mutant Collapsin Response Mediator Protein2 knock-in mice. Neurochem. Int. 2018, 119, 207–217. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, E.; Wang, Y.; Yang, C.; Yu, H.; Wang, Q.; Chen, Z.; Zhang, C.; Christian, K.M.; Song, H.; et al. Brain-specific Crmp2 deletion leads to neuronal development deficits and behavioural impairments in mice. Nat. Commun. 2016, 1, 7. [Google Scholar] [CrossRef]
- Miller, B.H.; Zeier, Z.; Xi, L.; Lanz, T.A.; Deng, S.; Strathmann, J.; Willoughby, D.; Kenny, P.J.; Elsworth, J.D.; Lawrence, M.S.; et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. USA 2012, 109, 3125–3130. [Google Scholar] [CrossRef] [Green Version]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef]
- Tsutiya, A.; Watanabe, H.; Nakano, Y.; Nishihara, M.; Goshima, Y.; Ohtani-Kaneko, R. Deletion of collapsin response mediator protein 4 results in abnormal layer thickness and elongation of mitral cell apical dendrites in the neonatal olfactory bulb. J. Anat. 2016, 228, 792–804. [Google Scholar] [CrossRef]
- Diagnostic and Statistical Manual of Mental Disorders, 5th ed (DSM-V); American Psychiatric Association: Philadelphia, PA, USA, 2013.
- Tsutiya, A.; Nishihara, M.; Goshima, Y.; Ohtani-Kaneko, R. Mouse pups lacking collapsin response mediator protein 4 manifest impaired olfactory function and hyperactivity in the olfactory bulb. Eur. J. Neurosci. 2015, 42, 2335–2345. [Google Scholar] [CrossRef]
- Takarae, Y.; Sablich, S.R.; White, S.P.; Sweeney, J.A. Neurophysiological hyperresponsivity to sensory input in autism spectrum disorders. J. Neurodev. Disord. 2016, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Takarae, Y.; Sweeney, J. Neural Hyperexcitability in Autism Spectrum Disorders. Brain Sci. 2017, 7, 129. [Google Scholar] [CrossRef]
- Spencer, C.M.; Alekseyenko, O.; Hamilton, S.M.; Thomas, A.M.; Serysheva, E.; Yuva-Paylor, L.A.; Paylor, R. Modifying behavioral phenotypes in Fmr1KO mice: Genetic background differences reveal autistic-like responses. Autism Res. 2011, 4, 40–56. [Google Scholar] [CrossRef]
- He, C.X.; Cantu, D.A.; Mantri, S.S.; Zeiger, W.A.; Goel, A.; Portera-Cailliau, C. Tactile defensiveness and impaired adaptation of neuronal activity in the Fmr1 knock-out mouse model of autism. J. Neurosci. 2017, 37, 6475–6487. [Google Scholar] [CrossRef]
- Ethridge, L.E.; White, S.P.; Mosconi, M.W.; Wang, J.; Byerly, M.J.; Sweeney, J.A. Reduced habituation of auditory evoked potentials indicate cortical hyper-excitability in Fragile X Syndrome. Transl. Psychiatry 2016, 6, e787. [Google Scholar] [CrossRef]
- Schmeisser, M.J.; Ey, E.; Wegener, S.; Bockmann, J.; Stempel, A.V.; Kuebler, A.; Janssen, A.L.; Udvardi, P.T.; Shiban, E.; Spilker, C.; et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 2012, 486, 256–260. [Google Scholar] [CrossRef]
- Won, H.; Lee, H.R.; Gee, H.Y.; Mah, W.; Kim, J.I.; Lee, J.; Ha, S.; Chung, C.; Jung, E.S.; Cho, Y.S.; et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 2012, 486, 261–265. [Google Scholar] [CrossRef]
- Ko, H.G.; Oh, S.B.; Zhuo, M.; Kaang, B.K. Reduced acute nociception and chronic pain in Shank2−/− mice. Mol. Pain 2016, 4, 12. [Google Scholar] [CrossRef]
- Scattoni, M.L.; Crawley, J.; Ricceri, L. Ultrasonic vocalizations: A tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci. Biobehav. Rev. 2009, 33, 508–515. [Google Scholar] [CrossRef]
- Van der Gucht, E.; Clerens, S.; Cromphout, K.; Vandesande, F.; Arckens, L. Differential expression of c-fos in subtypes of GABAergic cells following sensory stimulation in the cat primary visual cortex. Eur. J. Neurosci. 2002, 16, 1620–1626. [Google Scholar] [CrossRef]
- Sullivan, S.L.; Ressler, K.J.; Buck, L.B. Spatial patterning and information coding in the olfactory system. Curr. Opin. Genet. Dev. 1995, 5, 516–523. [Google Scholar] [CrossRef]
- Mombaerts, P.; Wang, F.; Dulac, C.; Chao, S.K.; Nemes, A.; Mendelsohn, M.; Edmondson, J.; Axel, R. Visualizing an olfactory sensory map. Cell 1996, 87, 675–686. [Google Scholar] [CrossRef]
- Salcedo, E.; Zhang, C.; Kronberg, E.; Restrepo, D. Analysis of training-induced changes in ethyl acetate odor maps using a new computational tool to map the glomerular layer of the olfactory bulb. Chem. Senses. 2005, 30, 615–626. [Google Scholar] [CrossRef]
- Pathania, M.; Davenport, E.C.; Muir, J.; Sheehan, D.F.; López-Doménech, G.; Kittler, J.T. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry. 2014, 4, e374. [Google Scholar] [CrossRef]
- Nagaoka, A.; Takehara, H.; Hayashi-Takagi, A.; Noguchi, J.; Ishii, K.; Shirai, F.; Yagishita, S.; Akagi, T.; Ichiki, T.; Kasai, H. Abnormal intrinsic dynamics of dendritic spines in a fragile X syndrome mouse model in vivo. Sci. Rep. 2016, 6, 26651. [Google Scholar] [CrossRef] [Green Version]
- Cheng, N.; Alshammari, F.; Hughes, E.; Khanbabaei, M.; Rho, J.M. Dendritic overgrowth and elevated ERK signaling during neonatal development in a mouse model of autism. PLoS ONE 2017, 12, e0179409. [Google Scholar] [CrossRef]
- Montani, C.; Ramon-Brossier, M.; Ponzoni, L.; Gritti, L.; Cwetsch, A.W.; Braida, D.; Saillour, Y.; Terragni, B.; Mantegazza, M.; Sala, M.; et al. The X-linked intellectual disability protein IL1RAPL1 regulates dendrite complexity. J. Neurosci. 2017, 37, 6606–6627. [Google Scholar] [CrossRef]
- Niisato, E.; Nagai, J.; Yamashita, N.; Abe, T.; Kiyonari, H.; Goshima, Y.; Ohshima, T. CRMP4 suppresses apical dendrite bifurcation of CA1 pyramidal neurons in the mouse hippocampus. Dev. Neurobiol. 2012, 72, 1447–1457. [Google Scholar] [CrossRef] [Green Version]
- Niisato, E.; Nagai, J.; Yamashita, N.; Nakamura, F.; Goshima, Y.; Ohshima, T. Phosphorylation of CRMP2 is involved in proper bifurcation of the apical dendrite of hippocampal CA1 pyramidal neurons. Dev. Neurobiol. 2013, 73, 142–151. [Google Scholar] [CrossRef]
- Cha, C.; Zhang, J.; Ji, Z.; Tan, M.; Li, S.; Wu, F.; Chen, K.; Gong, S.; Guo, G.; Lin, H. CRMP4 regulates dendritic growth and maturation via the interaction with actin cytoskeleton in cultured hippocampal neurons. Brain Res. Bull. 2016, 124, 286–294. [Google Scholar] [CrossRef]
- Takaya, R.; Nagai, J.; Piao, W.; Niisato, E.; Nakabayashi, T.; Yamazaki, Y.; Nakamura, F.; Yamashita, N.; Kolattukudy, P.; Goshima, Y.; et al. CRMP1 and CRMP4 are required for proper orientation of dendrites of cerebral pyramidal neurons in the developing mouse brain. Brain Res. 2017, 1655, 161–167. [Google Scholar] [CrossRef]
- Eissa, N.; Al-Houqani, M.; Sadeq, A.; Ojha, S.K.; Sasse, A.; Sadek, B. Current enlightenment about etiology and pharmacological treatment of autism spectrum disorder. Front. Neurosci. 2018, 12, 304. [Google Scholar] [CrossRef]
- Horder, J.; Petrinovic, M.M.; Mendez, M.A.; Bruns, A.; Takumi, T.; Spooren, W.; Barker, G.J.; Künnecke, B.; Murphy, D.G. Glutamate and GABA in autism spectrum disorder-a translational magnetic resonance spectroscopy study in man and rodent models. Transl. Psychiatry 2018, 8, 106. [Google Scholar] [CrossRef]
- Carlson, C.G. Glutamate receptor dysfunction and drug targets across models of autism spectrum disorders. Pharmacol. Biochem. Behav. 2012, 100, 850–854. [Google Scholar] [CrossRef]
- Kim, K.C.; Cho, K.S.; Yang, S.M.; Gonzales, E.L.; Valencia, S.; Eun, P.H.; Choi, C.S.; Mabunga, D.F.; Kim, J.W.; Noh, J.K.; et al. Sex differences in autism-like behavioral phenotypes and postsynaptic receptors expression in the prefrontal cortex of TERT transgenic mice. Biomol. Ther. 2017, 25, 374–382. [Google Scholar] [CrossRef]
- Fung, L.K.; Hardan, A.Y. Developing medications targeting glutamatergic dysfunction in autism: Progress to date. CNS Drugs 2015, 29, 453–463. [Google Scholar] [CrossRef]
- Silverman, J.L.; Tolu, S.S.; Barkan, C.L.; Crawley, J.N. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology 2010, 35, 976–989. [Google Scholar] [CrossRef]
- Mehta, M.V.; Gandal, M.J.; Siegel, S.J. mGluR5-antagonist mediated reversal of elevated stereotyped, repetitive behaviors in the VPA model of autism. PLoS ONE 2011, 6, e26077. [Google Scholar] [CrossRef]
- Gatto, C.L.; Broadie, K. Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front. Synaptic. Neurosci. 2010, 2, 4. [Google Scholar] [CrossRef]
- Rubenstein, J.L. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr. Opin. Neurol. 2010, 23, 118–123. [Google Scholar] [CrossRef]
- Jamain, S.; Betancur, C.; Quach, H.; Philippe, A.; Fellous, M.; Giros, B.; Gillberg, C.; Leboyer, M.; Bourgeron, T. Linkage and association of the glutamate receptor 6 gene with autism. Mol. Psychiatry 2002, 7, 302–310. [Google Scholar] [CrossRef] [Green Version]
- Naaijen, J.; Bralten, J.; Poelmans, G.; Glennon, J.C.; Franke, B.; Buitelaar, J.K. Glutamatergic and GABAergic gene sets in attention-deficit/hyperactivity disorder: Association to overlapping traits in ADHD and autism. Transl. Psychiatry. 2017, 7, e999. [Google Scholar] [CrossRef]
- Werling, D.M.; Geschwind, D.H. Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 2013, 26, 146–153. [Google Scholar] [CrossRef]
- Werling, D.M.; Geschwind, D.H. Recurrence rates provide evidence for sex-differential, familial genetic liability for autism spectrum disorders in multiplex families and twins. Mol. Autism. 2015, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Rubenstein, E.; Wiggins, L.D.; Lee, L.C. A review of the differences in developmental, psychiatric, and medical endophenotypes between males and females with autism spectrum disorder. J. Dev. Phys. Disabil. 2015, 27, 119–139. [Google Scholar] [CrossRef]
- Chen, C.; Van Horm, J.D. GENDAAR Research Consortium. Developmental neurogenetics and multimodal neuroimaging of sex differences in autism. Brain Imaging Behav. 2017, 11, 38–61. [Google Scholar] [CrossRef]
- Yu, J.; He, X.; Yao, D.; Li, Z.; Li, H.; Zhao, Z. A sex-specific association of common variants of neuroligin genes (NLGN3 and NLGN4X) with autism spectrum disorders in a Chinese Han cohort. Behav. Brain Funct. 2011, 7, 13. [Google Scholar] [CrossRef]
- Landini, M.; Merelli, I.; Raggi, M.E.; Galluccio, N.; Ciceri, F.; Bonfanti, A.; Camposeo, S.; Massagli, A.; Villa, L.; Salvi, E.; et al. Association Analysis of Noncoding Variants in Neuroligins 3 and 4X Genes with Autism Spectrum Disorder in an Italian Cohort. Int. J. Mol. Sci. 2016, 17, 1765. [Google Scholar] [CrossRef]
- Ey, E.; Torquet, N.; Le Sourd, A.M.; Leblond, C.S.; Boeckers, T.M.; Faure, P.; Bourgeron, T. The Autism ProSAP1/Shank2 mouse model displays quantitative and structural abnormalities in ultrasonic vocalisations. Behav. Brain Res. 2013, 256, 677–689. [Google Scholar] [CrossRef]
- Kim, K.C.; Kim, P.; Go, H.S.; Choi, C.S.; Park, J.H.; Kim, H.J.; Jeon, S.J.; Dela Pena, I.C.; Han, S.H.; Cheong, J.H.; et al. Male-specific alteration in excitatory post-synaptic development and social interaction in pre-natal valproic acid exposure model of autism spectrum disorder. J. Neurochem. 2013, 124, 832–843. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.; Roman, A.; Basta-Kaim, A.; Kubera, M.; Budziszewska, B.; Schneider, K.; Przewłockia, R. Gender-specific behavioral and immunological alterations in an animal model of autism induced by prenatal exposure to valproic acid. Psychoneuroendocrinology 2008, 33, 728–740. [Google Scholar] [CrossRef]
- Konopko, M.A.; Densmore, A.L.; Krueger, B.K. Sexually Dimorphic Epigenetic Regulation of Brain-Derived Neurotrophic Factor in Fetal Brain in the Valproic Acid Model of Autism Spectrum Disorder. Dev. Neurosci. 2017, 39, 507–518. [Google Scholar] [CrossRef]
- Nicolini, C.; Fahnestock, M. The valproic acid-induced rodent model of autism. Exp. Neurol. 2018, 299, 217–227. [Google Scholar] [CrossRef]
- Ju, A.; Hammerschmidt, K.; Tantra, M.; Krueger, D.; Brose, N.; Ehrenreich, H. Juvenile manifestation of ultrasound communication deficits in the neuroligin-4 null mutant mouse model of autism. Behav. Brain Res. 2014, 270, 159–164. [Google Scholar] [CrossRef]
- Iwakura, T.; Sakoh, M.; Tsutiya, A.; Yamashita, N.; Ohtani, A.; Tsuda, M.C.; Ogawa, S.; Tsukahara, S.; Nishihara, M.; Shiga, T.; et al. Collapsin response mediator protein 4 affects the number of tyrosine hydroxylase-immunoreactive neurons in the sexually dimorphic nucleus in female mice. Dev. Neurobiol. 2013, 73, 502–517. [Google Scholar] [CrossRef] [Green Version]
- Sumida, H.; Nishizuka, M.; Kano, Y.; Arai, Y. Sex differences in the anteroventral periventricular nucleus of the preoptic area and in the related effects of androgen in prenatal rats. Neurosci. Lett. 1993, 151, 41–44. [Google Scholar] [CrossRef]
- Ferri, S.L.; Abel, T.; Brodkin, E.S. Sex differences in autism spectrum disorder: A review. Curr. Psychiatry Rep. 2018, 20, 9. [Google Scholar] [CrossRef]
- Knickmeyer, R.C.; Baron-Cohen, S. Fetal testosterone and sex differences in typical social development and in autism. J. Child. Neurol. 2006, 21, 825–845. [Google Scholar] [CrossRef]
- Auyeung, B.; Baron-Cohen, S.; Ashwin, E.; Knickmeyer, R.; Taylor, K.; Hackett, G. Fetal testosterone and autistic traits. Br. J. Psychol. 2009, 100, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Auyeung, B.; Taylor, K.; Hackett, G.; Baron-Cohen, S. Foetal testosterone and autistic traits in 18 to 24-month-old children. Mol. Autism. 2010, 1, 11. [Google Scholar] [CrossRef]
- Auyeung, B.; Ahluwalia, J.; Thomson, L.; Taylor, K.; Hackett, G.; O’Donnell, K.J.; Baron-Cohen, S. Prenatal versus postnatal sex steroid hormone effects on autistic traits in children at 18 to 24 months of age. Mol. Autism. 2012, 3, 17. [Google Scholar] [CrossRef]
- Baron-Cohen, S.; Auyeung, B.; Nørgaard-Pedersen, B.; Hougaard, D.M.; Abdallah, M.W.; Melgaard, L.; Cohen, A.S.; Chakrabarti, B.; Ruta, L.; Lombardo, M.V. Elevated fetal steroidogenic activity in autism. Mol. Psychiatry 2015, 20, 369–376. [Google Scholar] [CrossRef]
- Cherskov, A.; Pohl, A.; Allison, C.; Zhang, H.; Payne, R.A.; Baron-Cohen, S. Polycystic ovary syndrome and autism: A test of the prenatal sex steroid theory. Transl. Psychiatry 2018, 8, 136. [Google Scholar] [CrossRef]
- Mong, J.A.; Glaser, E.; McCarthy, M.M. Gonadal steroids promote glial differentiation and alter neuronal morphology in the developing hypothalamus in a regionally specific manner. J. Neurosci. 1999, 19, 1464–1472. [Google Scholar] [CrossRef]
- McCarthy, M.M.; Wright, C.L. Convergence of Sex Differences and the Neuroimmune System in Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 402–410. [Google Scholar] [CrossRef]
- Werling, D.M.; Parikshak, N.N.; Geschwind, D.H. Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat. Commun. 2016, 7, 10717. [Google Scholar] [CrossRef] [Green Version]


© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohtani-Kaneko, R. Crmp4-KO Mice as an Animal Model for Investigating Certain Phenotypes of Autism Spectrum Disorders. Int. J. Mol. Sci. 2019, 20, 2485. https://doi.org/10.3390/ijms20102485
Ohtani-Kaneko R. Crmp4-KO Mice as an Animal Model for Investigating Certain Phenotypes of Autism Spectrum Disorders. International Journal of Molecular Sciences. 2019; 20(10):2485. https://doi.org/10.3390/ijms20102485
Chicago/Turabian StyleOhtani-Kaneko, Ritsuko. 2019. "Crmp4-KO Mice as an Animal Model for Investigating Certain Phenotypes of Autism Spectrum Disorders" International Journal of Molecular Sciences 20, no. 10: 2485. https://doi.org/10.3390/ijms20102485
APA StyleOhtani-Kaneko, R. (2019). Crmp4-KO Mice as an Animal Model for Investigating Certain Phenotypes of Autism Spectrum Disorders. International Journal of Molecular Sciences, 20(10), 2485. https://doi.org/10.3390/ijms20102485