1. Introduction
Durum (
Triticum turgidum L. subsp.
durum, genome AABB, 2n = 4× = 28) and common (
Triticum aestivum L. subsp.
aestivum, genome AABBDD, 2n = 6× = 42) wheats are species with significant agricultural importance as cereal grains throughout much of the world. In particular, durum wheat is grown mainly in the Mediterranean basin, and is primarily used for pasta, couscous and bread. Durum wheat, which originated in the Fertile Crescent, spread west of the Mediterranean basin, reaching the Iberian Peninsula and North Africa ca. 7000 years BC [
1,
2]. Wheat reached this area through Egypt, which acted as the bridge to the Fertile Crescent for the diffusion of wheat in North Africa. Other sources of wheat were maritime: wheat was introduced to Libya from Greece and Crete [
3]. Moreover from the south of the Italian peninsula through the Sicily region, wheat reached the coasts of Tunisia, Morocco and Algeria [
3]. This migration process, combined with both natural and human selection, led to the creation of local varieties along the Mediterranean basin that were well adapted to its different agro-ecological zones. Indeed, Tunisia and Algeria have a large collection of durum local varieties with great diversity, as observed for the first time by Boeuf in 1932 [
4]. Contrasting environmental conditions of North Africa triggered a wide diversification of the durum wheat genome determining the development of several local populations. Thereby, North Africa is considered one of the principal secondary centers of diversification of durum wheat [
5]. Studies on the origin and migration of durum wheat based on genetic diversity of landraces have shown that Tunisian landraces and those from the Mediterranean basin have a shared allelic identity and, consequently, that they originated from related seed sources and were exposed to similar environmental pressures and accumulated distinct mutations over time [
6]. Durum wheat cultivation has continued to the present day as a staple food for local populations and possesses an outstanding socio-economical value as an important basic constituent of the diet in Maghreb countries [
7,
8].
In Tunisia during the 2018/2019 season, 0.5 M ha of durum wheat were cultivated, representing about 85% of the area devoted to wheat (common and durum), which in turn accounted for more than half of the cereal cultivated area [
9]. The durum wheat germplasm cultivated includes about 45 varieties [
10] grouped in (a) traditional varieties selected from Tunisian landraces (developed up to 1930s), (b) old cultivars produced by crossing Tunisian traditional varieties and foreign materials (developed up to 1970s) and (c) modern cultivars obtained in national and international breeding centers (developed up to present). The traditional varieties still prevail in rural areas where farmers still use traditional farming systems due to the impossibility of adopting modern intensive ones, or where this germplasm ensures tolerance to stresses [
11,
12]. The traditional varieties can be defined as the consequence of an empiric breeding program focused on exploiting the phenotypic variability among Tunisian durum landraces, such as ‘Biskri’, ‘Bidi’, ‘Mahmoudi’, ‘Chili’ and ‘Jenah Khottifa’ [
7,
8].
Several descriptions exist to define a ‘landrace’ [
13,
14]. Villa et al. [
15] remarked that a landrace is ‘a dynamic population of a cultivated plant that has historical origin, distinct identity and lacks formal crop improvement, as well as often being genetically diverse, locally adapted and associated with traditional farming systems’. An autochthonous wheat landrace can be defined as a traditional variety with a high capacity to tolerate biotic and abiotic stresses, resulting in high yield stability and an intermediate yield level under a low input agricultural system [
16]. Wheat landraces are valuable sources for broadening the genetic base of cultivated wheat. In fact, several studies using morphological, physiological and agronomic traits have shown that genetically diverse local germplasm well adapted to a broad range of environmental conditions can be considered an important reservoir of useful genes for exploitation in wheat breeding programs [
17,
18,
19,
20].
Molecular markers provide a satisfactory option for several genetic plant studies, since they are highly polymorphic and almost unlimited in number, do not vary among tissues or developmental stages of a plant and are not affected by environmental factors. Therefore, molecular markers can be used for genome analysis, and many of these have been applied with success for genetic mapping [
21,
22], cultivar identification [
23] and diversity studies [
19,
20]. Different molecular markers have been developed (see review [
24]). In particular, simple sequence repeats (SSRs) markers represent a useful alternative marker system because of their abundance in the wheat genome and easy detection as PCR-based molecular markers. Moreover, the advantages of SSRs include high information content, codominant inheritance, locus specificity and high reproducibility. Fifteen SSRs and five primer pair combinations of amplified fragment length polymorphism (AFLP) were tested by Medini et al. [
10] in a wheat collection including Tunisian cultivars and wild wheat species. Cluster analysis separated the wild wheat species from the durum wheat cultivars, and these latter were further split in old and modern cultivars. The discriminant analysis carried out on 196 durum wheat lines issued from landraces collected in Tunisia and genotyped with diversity array technology sequencing (DArTseq) distinguished five groups that matched with the farmer’s variety nomenclature [
25].
Nowadays, thanks to the activity of the National Gene Bank of Tunisia (NGBT), a program to collect durum wheat germplasm has been started. This program includes the idea of fostering the cultivation of traditional Tunisian durum germplasm by on farm conservation [
26]. Within this framework, the maintenance and sustainable management of the genetic diversity present in autochthonous durum wheats are an absolute priority in order to preserve this genetic heritage, ensure food sovereignty and help smallholders cope with climate change. In this context the main goals of this work were to: (i) explore the genetic diversity in Tunisian durum wheat germplasm collected in different agro-ecological regions; (ii) evaluate the genetic structure of the Tunisian durum wheat genotypes; and (iii) clarify synonymy cases among the Tunisian durum wheat samples collected.
3. Discussion
During the second half of the last century, a significant increase in wheat yield was observed in Tunisia. This was a consequence of the introduction of modern high-yielding varieties able to optimize water and soil resources, as well as a result of improvements in agronomic techniques, such as better crop management [
7,
8].
In Tunisia, as in the whole Mediterranean region, rainfall is highly variable in its yearly distribution [
27]. The overall yearly rainfall is differently distributed over the country, with northern regions much wetter than the Center and South (
Figure 1 and
Figure S1). The growing season of wheat, with sowing between November and December, extends through the wet and cool season when in central/southern regions there is a reduced level of precipitation but evaporation is relatively low, thus securing enough water for the plants’ needs. More critical is the period between April and May, the period of flowering-to-grain filling, when high temperatures or drought stress can dramatically reduce kernel setting, and thus yield [
22,
27,
31]. In this period, the difference in precipitation among the three regions of Tunisia drastically increases (
Figure S1). Therefore, it is possible to suppose that plants adapted to central and principally southern environments should possess traits of adaptation, allowing them to escape from these limiting environmental conditions.
In our study, a set of 16 SSR markers with a good coverage of the A and B genomes of durum wheat were tested in the composed durum wheat collection. They allowed the identification of 95 alleles, with an average of 5,9 alleles/SSR locus. The considerable number of alleles was due to the multi-allelic nature of these markers that facilitates variability detection [
32]. Similar results were reported by Chen et al. [
30] in a Chinese winter wheat collection, although a large range of alleles per locus (4.5–18.1) was reported in the literature for a wider germplasm collection [
33,
34]. The genetic diversity of the collection (
He = 0.58) was similar to the values reported by previous studies involving durum wheat collections (
He values between 0.55 and 0.68) [
33,
35].
To define the genetic distances among samples, the matrix obtained with a simple matching coefficient was also used to produce a neighbor-joining dendrogram (
Figure 2). This dendrogram clearly divided collected germplasm into two distinct groups: the one hosting only traditional varieties, (mostly Biskri, Mahmoudi and Chili), and one with a more assorted composition (including both traditional and modern varieties). In this latter group, the distances separating the entries are much higher than in the former one, which we will refer to as the ‘core traditional varieties’ (CTVs) group.
To better investigate this result, an analysis of the molecular variance was performed to test different sources of variation. In the first instance, we tested the distribution of molecular variation among the CTV group and the rest of the analyzed germplasm. The results clearly indicate that the CTV group is unquestionably differentiated from the rest of the analyzed material, since 22% of the total variation is between the two groups of samples (
Table 2). In a second run, three groups were tested: the CTV, the one formed by entries described by farmers as traditional varieties and, finally, modern cultivars. In this case, AMOVA demonstrated that the distribution of the molecular variance conformed to the grouping, even though at a lower extent than in the former analysis (
Table S1).
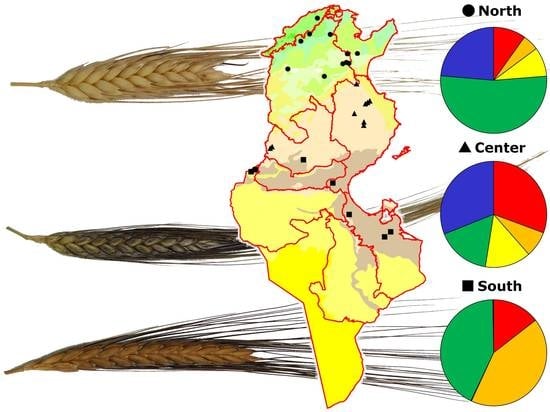
Nevertheless, the above analyses did not provide any information on the distribution of genotypes along the geographic cline. To this end a Bayesian approach was utilized by analyzing genotypic data through the STRUCTURE software. Structure analysis assigns each genotype to a specific subpopulation and five different ones were identified after the Evanno test [
29]. The robustness of these results was confirmed by the AMOVA used to test the distribution of variation among and within these five subpopulations. In fact, the AMOVA indicated that 37% of the total variation observed was among these five groups (
Table 3). After assigning each genotype to a specific subpopulation, as described in the results section, we tested the richness of each subpopulation using the three collection zones (North, Center, and South) as discriminating factors for Tunisian samples; a fourth discriminating factor was the modern durum cultivars (see
Figure 3).
The approach described above allowed us to observe that some genotypes (belonging to the subpopulations indicated with the colors red and orange in
Figure 3) were increasingly frequent along the North–South cline, while at the same time the modern cultivars tended to be decreasingly represented. Conversely, in the modern cultivars, the presence of traditional germplasm was scarce.
This picture indicates that: (a) modern varieties are more fit for cultivation in the northern part of Tunisia, where the rainfall is adequate for new high-yielding varieties that develop their full potential only in the presence of moderate to high inputs; (b) there is an increased presence of traditional varieties and CTVs in the Center and South groups, which are characterized by reduced and more erratic precipitations.
These findings indicate that the traditional varieties, originally developed from North African landraces, might possess traits of resistance to unfavorable climatic conditions that are absent in the modern cultivars. This is not surprising since landraces and derived traditional varieties are the result of a non-scientific process of selection driven by farmers’ cultural values and by natural adaptation to local environment [
13].
The fact that some old cultivars, such as AgiliAC2, are still called by the names of traditional varieties might be the result of different causes. In fact, the improved material of the 1970s–1990s might have been distributed using the names of the traditional varieties in order to overcome the diffidence of more traditional farmers against innovations. Another reason might reside in a phenotypic resemblance of some morphological traits of the new introductions with those of the CTVs, or by the misnaming of landraces during seed exchange among farmers or during the collecting missions undertaken by the NGBT [
25].
In conclusion, our results showed that the molecular markers distinguished Tunisian durum wheat germplasm (named the CTV group) and detected putative synonymy and homonymy cases. These results could be useful to increase ex situ conservation efficiency, because SSRs allow the detection of redundant duplications within durum germplasm conserved in the NGBT. In situ on-farm conservation, in addition to the ex situ approach, could ensure the long-term conservation of genetically diverse Tunisian durum wheat available in local germplasm, which is a reservoir of genes for drought, heat and salinity tolerance and quality traits. The traditional varieties could help maintain the use of local knowledge and challenge ongoing genetic erosion due to the spread of modern pure line cultivars.
The population structure analysis suggests a strong genetic stratification along the North–South cline of Tunisia. This could be the result of breeding programs from the second half of the last century that produced high-yielding varieties for the northern highly productive areas rather than tackling the agro-ecological conditions of the Center and South.
The strategy of incorporating adaptation traits found in the genetic diversity pool of landraces and traditional varieties into modern breeding programs is widely supported [
36]. In this way it will also be possible to contend with the challenges imposed by climate change and the survival of agriculture in marginal lands [
36,
37,
38]. Correct and scientifically driven programs aiming at
in situ conservation of agro-biodiversity on farms are essential, particularly for those species that are the food staple of Mediterranean populations and Tunisians in particular, such as durum wheat [
39]. In fact, the end-use caracteristics are among the selection drivers used by past farmers during the non-scientific selection of traditional landraces. This explains the preference of genotypes providing better couscous or more valuable tabouna (a traditional sort of bread).