AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis
Abstract
:1. Introduction
2. Results
2.1. Hypocotyl Hook Bending Angle Differences between Col-0 and Atcrk5-1
2.2. Kinetics of GA3-Regulated Apical Hook Development
2.3. AtCRK5 is a Regulator of the Auxin Maxima in the Apical Hook
2.3.1. Distribution of the Auxin in Hypocotyl Hooks monitored by DR5::GFP Fluorescence
2.3.2. Distribution of PIN3-GFP, PIN7-GFP and AUX1-YFP in Hypocotyl Hooks
2.4. Gene Expression Studies by qRT-PCR
2.4.1. Expression of the Auxin Biosynthesis and Catabolism Genes without/with ACC Treatment
2.4.2. Expression of the Ethylene Biosynthesis and Signaling Genes without/with ACC Treatment
2.4.3. Expression of the PAT Genes without/with ACC Treatment
2.5. AtCRK5 Can Phosphorylate the Auxin Efflux PIN3 Protein in vitro
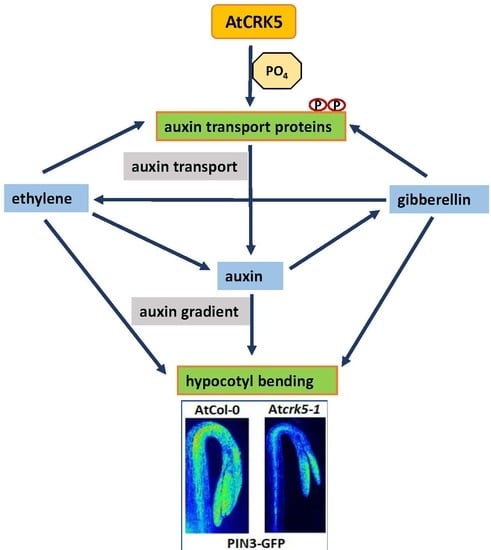
3. Discussion
3.1. AtCRK5 Might be a General Regulator of PIN Proteins, Auxin Distribution, and Differential Organ Growth
3.2. The AtCRK5 Kinase Influences the Hormonal Crosstalk Regulating Hypocotyl Hook Development
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Time Lapse Assays
4.3. Total GA Measurement by Competitive GAs Elisa Assay
4.4. RNA Isolation and Real Time Quantitative PCR (qRT PCR) for Hypocotyl Gene Expression
4.5. PINs-GFP Protein Abundance Monitoring in Hypocotyl Hooks by LSM Microscopy
4.6. In vitro Kinase Assay
4.6.1. PIN hydrophilic Loop Region Cloning
4.6.2. Purification of 6XHis Tagged Protein
4.6.3. In vitro Kinase Assays
4.7. Bioinformatics Analysis
4.8. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vandenbussche, F.; Verbelen, J.P.; Van Der Straeten, D. Of light and length: Regulation of hypocotyl growth in Arabidopsis. Bioessays 2005, 27, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Shi, H.; Xue, C.; Wei, N.; Guo, H.; Deng, X.W. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 2014, 111, 3913–3920. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhang, Y.; Wang, J.; Yan, X.; Wang, C.; Xu, J.; Pan, J. Clathrin-Mediated Auxin Efflux and Maxima Regulate Hypocotyl Hook Formation and Light-Stimulated Hook Opening in Arabidopsis. Mol. Plant 2016, 9, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salome, P.A. Life’s a gas under pressure: Ethylene and etioplast maintenance in germinating seedlings. Plant Cell 2017, 29, 2951–2952. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Petrásek, J.; Zádníková, P.; Hoyerová, K.; Pesek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zazímalová, E.; Benková, E.; et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Zadnikova, P.; Petrasek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J.; Arana, M.V.; Vandenbussche, F.; Zadnikova, P.; Minguet, E.G.; Guardiola, V.; Van Der Straeten, D.; Benkova, E.; Alabadi, D.; Blazquez, M.A. Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 2011, 67, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Raz, V.; Ecker, J.R. Regulation of differential growth in the apical hook of Arabidopsis. Development 1999, 126, 3661–3668. [Google Scholar] [PubMed]
- Li, H.; Johnson, P.; Stepanova, A.; Alonso, J.M.; Ecker, J.R. Convergence of signaling of differential cell growth pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 2004, 7, 193–204. [Google Scholar] [CrossRef]
- Abbas, M.; Alabadi, D.; Blazquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441–450. [Google Scholar] [CrossRef]
- Mazzella, M.A.; Casal, J.J.; Muschietti, J.P.; Fox, A.R. Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 2014, 5, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Zadnikova, P.; Smet, D.; Zhu, Q.; Van Der Straeten, V.; Benkova, E. Strategies of seedlings to overcome their sessile nature: Auxin in mobility control. Front. Plant Sci. 2015, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Zádnikova, P.; Wabnik, K.; Abuzeineh, A.; Gallemi, M.; Van Der Straeten, D.; Smith, R.S.; Inze, D.; Friml, J.; Prusinkiewicz, P.; Benkova, E. A model of differential growth-guided apical hook formation in plants. Plant Cell 2016, 28, 2464–2477. [Google Scholar]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Petrasek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertova, D.; Wiśniewska, J.; Tadele, Z.; Kubeš, M.; Čovanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Grunewald, W.; Sauer, M.; Cannoot, B.; Soriano, M.; Swarup, R.; Weijers, D.; Bennett, M.; Boutilier, K.; Friml, J. Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 2015, 142, 702–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dong, Q.; Kita, D.; Huang, J.B.; Liu, G.; Wu, X.; Zhu, X.; Cheung, A.Y.; Wu, H.M.; Tao, L.Z. RopGEF1 Plays a Critical Role in Polar Auxin Transport in Early Development. Plant Physiol. 2017, 175, 157–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.J.; Luo, J. The PIN-FORMED auxin efflux carriers in plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzmán, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523. [Google Scholar]
- Roman, G.; Lubarsky, B.; Kieber, J.J.; Rothenberg, M.; Ecker, J.R. Genetic Analysis of Ethylene Signal Transduction in Arabidopsis thaliana: Five Novel Mutant Loci Integrated into a Stress Response Pathway. Genetics 1995, 139, 1393–1409. [Google Scholar] [PubMed]
- Lehman, A.; Black, R.; Ecker, J.R. (1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 1996, 85, 183–194. [Google Scholar] [CrossRef]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front. Plant Sci. 2014, 5. Article412/1. [Google Scholar] [CrossRef] [PubMed]
- Daviere, J.M.; Achard, P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Ogiso-Tanaka, E.; Zourelidou, M.; Schwechheimer, C. WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 2012, 139, 4020–4028. [Google Scholar] [CrossRef] [Green Version]
- Frigerio, M.; Alabadí, D.; Pérez-Gómez, J.; García-Cárcel, L.; Phillips, A.L.; Hedden, P.; Blázquez, M.A. Transcriptional Regulation of Gibberellin Metabolism Genes by Auxin Signaling in Arabidopsis. Plant Physiol. 2006, 142, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.; Estelle, M. Hypocotyl Transcriptome Reveals Auxin Regulation of Growth-Promoting Genes through GA-Dependent and -Independent Pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Annual Plant Reviews, The Gibberellins, 1st ed.; Wiley and Sons, Ltd.: Chichester, UK, 2016; pp. 285–312. [Google Scholar]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Xu, Y.P.; Munyampundu, J.P.; Liu, T.Y.; Cai, X.Z. Calcium dependent protein kinase (CDPK) and CDPK related kinase (CRK) gene families in tomato: Genome wide identification and functional analyses indisease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Rigó, G.; Andrási, N.; Tietz, O.; Palme, K.; Szabados, L.; Cséplő, Á. Striving Towards Abiotic Stresses: Role of the Plant CDPK Superfamily Members. In International Climate Protection; Palocz-Andresen, M., Szalay, D., Gosztom, A., Sípos, L., Taligás, T., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 99–105. [Google Scholar]
- Renna, L.; Stefano, G.; Majeran, W.; Micalella, C.; Meinnel, T.; Giglione, C.; Brandizzi, F. Golgi traffic integrity depends on N-Myristoyl transferase-1 in Arabidopsis. Plant Cell 2013, 25, 1756–1773. [Google Scholar] [CrossRef]
- Xu, W.; Huang, W. Calcium-Dependent Protein Kinases in Phytohormon Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 2436. [Google Scholar] [CrossRef]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium Dependent Protein Kinase, a Versatile Player in Plant Stress Management and Development. Crit. Rev. Plant Sci. 2017, 36, 336–352. [Google Scholar] [CrossRef]
- Rigó, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovács, H.; Páy, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef]
- Nemoto, K.; Takemori, N.; Seki, M.; Shinozaki, K.; Sawasaki, T. Members of the Plant CRK Superfamily Are Capable of Trans- and Autophosphorylation of Tyrosine Residues. J. Biol. Chem. 2015, 290, 16665–16677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takanoa, J.; Tanaka, M.; Toyoda, A.; Miwa, K.; Kasai, K.; Fuji, K.; Onouchi, H.; Naito, S.; Fujiwara, T. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 5220–5225. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.Y.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Ann. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Ottenschläger, I.; Wolff, P.; Wolverton, C.; Bhalerao, R.P.; Sandberg, G.; Ishikawa, H.; Evans, M.; Palme, K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 2003, 100, 2987–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baster, P.; Robert, S.; Kleine-Vehn, J.; Vanneste, S.; Kania, U.; Grunewald, W.; De Rybel, B.; Beeckman, T.; Friml, J. SCFTIR1/AFB-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 2013, 32, 260–274. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Friml, J.; Marchant, A.; Ljung, K.; Sandberg, G.; Klaus Palme, K.; Malcolm Bennett, M. Localization of the auxin permease AUX 1suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001, 15, 2648–2653. [Google Scholar] [CrossRef]
- Béziat, C.; Kleine-Vehn, J. The Road to Auxin-Dependent Growth Repression and Promotion in Apical Hooks. Curr. Biol. 2018, 28, R519–R525. [Google Scholar] [CrossRef] [Green Version]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249–260. [Google Scholar] [CrossRef]
- Abas, L.; Benjamins, R.; Malenica, N.; Paciorek, T.; Wiśniewska, J.; Moulinier–Anzola, J.C.; Sieberer, T.; Friml, J.; Luschnig, C. Intracellular trafficking and proteolysis of the Arabidopsis auxin efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 2006, 8, 249–256. [Google Scholar] [CrossRef]
- Kleine-Vehn, J.; Leitner, J.; Zwiewka, M.; Sauer, M.; Abas, L.; Luschnig, C.; Friml, J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 2008, 105/46, 17812–17817. [Google Scholar] [CrossRef] [PubMed]
- Leitner, J.; Petrásek, J.; Tomanov, K.; Retzer, K.; Parezová, M.; Korbei, B.; Bachmair, A.; Zažímalová, E.; Luschnig, C. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Prot Natl. Acad. Sci. USA 2012, 109/21, 8322–8327. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010, 62, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Koczka, L. Determination of the Arabidopsis thaliana CRK5 protein kinase phosphorylation sites on the PIN1, PIN2 and PIN3 hydrophilic loop region. Master’s Thesis, University of Szeged, Szeged, Hungary, 2016. [Google Scholar]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Dhonukshe, P.; Huang, F.; Galvan-Ampudia, C.S.; Mähönen, A.P.; Kleine-Vehn, J.; Xu, J.; Quint, A.; Prasad, K.; Friml, J.; Scheres, B.; et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 2010, 137, 3245–3255. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Nodzynski, T.; Pencík, A.; Rolcík, J.; Friml, J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 2010, 107, 918–922. [Google Scholar] [CrossRef]
- Ganguly, A.; Lee, S.H.; Cho, H.T. Functional identification of the phosphorylation sites of Arabidopsis PIN-FORMED3 for its subcellular localization and biological role. Plant J. 2012, 71, 810–823. [Google Scholar] [CrossRef]
- Barbosa, I.C.R.; Hammes, U.Z.; Schwechheimer, C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018, 23/6, 523–538. [Google Scholar] [CrossRef]
- Schwechheimer, C. Gibberellin signaling in plants—The extended version. Front. Plant Sci. 2012, 2. 107. [Google Scholar] [CrossRef]
- Salanenka, Y.; Verstraeten, I.; Löfke, C.; Tabata, K.; Naramoto, S.; Glanc, M.; Friml, J. Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc. Natl. Acad. Sci. USA 2018, 115/14, 3716–3721. [Google Scholar] [CrossRef]
- Sun, T.-P.; Kamiya, Y. The Arabidopsis GAl Locus Encodes the Cyclase ent-Kaurene Synthetase A of Gibberellin Biosynthesis. Plant Cell 1994, 6, 1509–1518. [Google Scholar] [PubMed]
- Willige, C.V.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.N.; Maier, A.; Schwechheimer, C. The DELLA Domain of GA INSENSITIVE Mediates the Interaction with the GA INSENSITIVE DWARF1A Gibberellin Receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Isono, E.; Richter, R.; Zourelidou, M.; Schwechheimer, C. Gibberellin Regulates PIN-FORMED Abundance and Is Required for Auxin Transport–Dependent Growth and Development in Arabidopsis thaliana. Plant Cell 2011, 23, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, N.; Ellis, J.; Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R.Acad.Sci. Paris Life Sci. 1993, 316, 1194–1199. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baba, A.I.; Andrási, N.; Valkai, I.; Gorcsa, T.; Koczka, L.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; Fehér, A.; Rigó, G.; et al. AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis. Int. J. Mol. Sci. 2019, 20, 3432. https://doi.org/10.3390/ijms20143432
Baba AI, Andrási N, Valkai I, Gorcsa T, Koczka L, Darula Z, Medzihradszky KF, Szabados L, Fehér A, Rigó G, et al. AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis. International Journal of Molecular Sciences. 2019; 20(14):3432. https://doi.org/10.3390/ijms20143432
Chicago/Turabian StyleBaba, Abu Imran, Norbert Andrási, Ildikó Valkai, Teréz Gorcsa, Lilla Koczka, Zsuzsanna Darula, Katalin F. Medzihradszky, László Szabados, Attila Fehér, Gábor Rigó, and et al. 2019. "AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis" International Journal of Molecular Sciences 20, no. 14: 3432. https://doi.org/10.3390/ijms20143432
APA StyleBaba, A. I., Andrási, N., Valkai, I., Gorcsa, T., Koczka, L., Darula, Z., Medzihradszky, K. F., Szabados, L., Fehér, A., Rigó, G., & Cséplő, Á. (2019). AtCRK5 Protein Kinase Exhibits a Regulatory Role in Hypocotyl Hook Development during Skotomorphogenesis. International Journal of Molecular Sciences, 20(14), 3432. https://doi.org/10.3390/ijms20143432