Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum
Abstract
:1. Introduction
2. Results
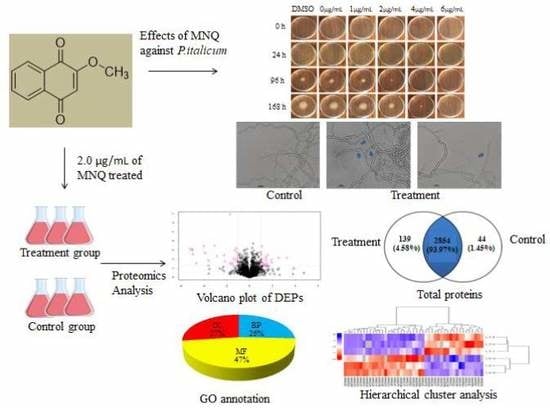
2.1. Effects of MNQ on P. italicum
2.2. Light Microscopy Observation
2.3. Label-Free Proteomic Based Protein Identification in the Treatment Group and Control Group
2.4. Functional Annotation of Proteins
2.4.1. Gene Ontology (GO) Annotation
2.4.2. Metabolic Pathway Annotation
2.5. PRM Verification of DEPs
3. Discussion
3.1. Effects of MNQ on the Mycelial Growth and Morphology of P. italicum
3.2. Proteins Related to Energy Generation and the NADPH Supply and Other Functions
4. Materials and Methods
4.1. Microorganism and Medium
4.2. MNQ Inhibition of P. italicum
4.2.1. Antifungal Assay
4.2.2. Minimum Inhibitory Concentration (MIC) Tests
4.3. Cell Culture and MNQ Treatment
4.4. Label-Free Quantitative Proteomics
4.4.1. Sample Preparation
4.4.2. Protein Digestion
4.4.3. Liquid Chromatography (LC)-Electrospray Ionization (ESI) Tandem MS (MS/MS) Analysis
4.4.4. Sequence Database Searching and Data Analysis
4.5. Parallel Reaction Monitoring (PRM) Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MNQ | 2-Methoxy-1,4-naphthoquinone |
| MIC | Minimum inhibitory concentration |
| PRM | Parallel reaction monitoring |
| FDR | False discovery rate |
| DEPs | Differentially expressed proteins |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
| ACAD | Acyl-CoA dehydrogenase |
| ME | Malic enzyme |
| 6PGDH | 6-Phosphogluconate dehydrogenase |
| ROS | Reactive oxygen species |
| CAT | Catalase |
| GS | Glutathione synthetase |
| PPP | Pentose phosphate pathway |
| RPE | Ribulose-phosphate 3-epimerase |
| KOs | Protein number at KEGG database |
| PDA | Potato dextrose agar |
| PDB | Potato dextrose broth |
| HCD | Higher energy dissociation |
| pAGC | Predictive automatic gain control |
| DMSO | Dimethylsulfoxide |
| PBS | Phosphate buffered saline |
References
- Rabea, E.I.; Badawy, M.E.I.; Steurbaut, W.; Stevens, C.V. In vitro assessment of N-(benzyl)chitosan derivatives against some plant pathogenic bacteria and fungi. Eur. Polym. J. 2009, 45, 237–245. [Google Scholar] [CrossRef]
- Gao, Y.Y.; He, L.F.; Li, X.X.; Lin, J.; Mu, W.; Liu, F. Toxicity and biochemical action of the antibiotic fungicide tetramycin on Colletotrichum scovillei. Pestic. Biochem. Phys. 2018, 147, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Qi, W.W.; Peng, X.; Chen, J.Y.; Wan, C.P. Inhibitory effect of 7-demethoxytylophorine on Penicillium italicum and its possible mechanism. Microorganisms 2019, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Talibi, I.; Boubaker, H.; Boudyach, E.H.; Ben Aoumar, A.A. Alternative methods for the control of postharvest citrus diseases. J. Appl. Microbiol. 2014, 117, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.P.; Li, P.; Chen, C.Y.; Peng, X.; Li, M.X.; Chen, M.; Wang, J.S.; Chen, J.Y. Antifungal activity of Ramulus cinnamomi explored by 1H-NMR based metabolomics approach. Molecules 2017, 22, 2237. [Google Scholar] [CrossRef] [PubMed]
- Tao, N.G.; Jia, L.; Zhou, H.E.; He, X.L. Effect of octanal on the mycelial growth of Penicillium italicum and P-digitatum. World J. Microb. Biot. 2014, 30, 1169–1175. [Google Scholar] [CrossRef]
- Tao, N.G.; OuYang, Q.L.; Jia, L. Citral inhibits mycelial growth of Penicillium italicum by a membrane damage mechanism. Food Control 2014, 41, 116–121. [Google Scholar] [CrossRef]
- Yang, S.Z.; Zhou, J.; Li, D.M.; Shang, C.Y.; Peng, L.T.; Pan, S.Y. The structure-antifungal activity relationship of 5,7-dihydroxyflavonoids against Penicillium italicum. Food Chem. 2017, 224, 26–31. [Google Scholar] [CrossRef]
- Jiang, H.F.; Zhuang, Z.H.; Hou, B.W.; Shi, B.J.; Shu, C.J.; Chen, L.; Shi, G.X.; Zhang, W.M. Adverse effects of hydroalcoholic extracts and the major components in the stems of Impatiens balsamina L. on Caenorhabditis elegans. Evid. Based Complement. Altern. Med. 2017, 2017, 4245830. [Google Scholar] [CrossRef]
- Wang, Y.C.; Lin, Y.H. Anti-gastric adenocarcinoma activity of 2-methoxy-1,4-naphthoquinone, an anti-Helicobacter pylori compound from Impatiens balsamina L. Fitoterapia 2012, 83, 1336–1344. [Google Scholar] [CrossRef]
- Liew, K.; Yong, P.V.C.; Navaratnam, V.; Lim, Y.M.; Ho, A.S.H. Differential proteomic analysis on the effects of 2-methoxy-1,4-naphthoquinone towards MDA-MB-231 cell line. Phytomedicine 2015, 22, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Colwell, C.A.; Mccall, M. Studies on the mechanism of antibacterial action of 2-methyl-1,4-naphthoquinone. Science 1945, 101, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.S.; Jiang, F.S.; Chen, N.P.; Lv, G.Y.; Zhu, C.G. Isolation and identification of an anti-tumor component from leaves of Impatiens balsamina. Molecules 2008, 13, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.C.; Zou, J.; Wang, G.D. Advances in the proteomic discovery of novel therapeutic targets in cancer. Drug Des. Dev. Ther. 2013, 7, 1259–1271. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Pai, P.J.; Zhang, W.P.; Hu, Y.W.; Dong, X.J.; Qian, P.Y.; Chen, D.J.; Lam, H. Proteomic response of methicillin-resistant S. aureus to a synergistic antibacterial drug combination: A novel erythromycin derivative and oxacillin. Sci. Rep. 2016, 6, 19841. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Liu, H.F.; Lin, Y.Z.; Wang, X.F.; Ma, J.; Li, Y.J.; Wang, X.J.; Zhou, J.H. Proteomic alteration of equine monocyte-derived macrophages infected with equine infectious anemia virus. Proteomics 2015, 15, 1843–1858. [Google Scholar] [CrossRef]
- Wenzel, M.; Bandow, J.E. Proteomic signatures in antibiotic research. Proteomics 2011, 11, 3256–3268. [Google Scholar] [CrossRef]
- Burchmore, R. Mapping pathways to drug resistance with proteomics. Expert Rev. Proteom. 2014, 11, 1–3. [Google Scholar] [CrossRef]
- Parveen, M.; Hasan, M.K.; Takahashi, J.; Murata, Y.; Kitagawa, E.; Kodama, O.; Iwahashi, H. Response of Saccharomyces cerevisiae to a monoterpene: Evaluation of antifungal potential by DNA microarray analysis. J. Antimicrob. Chemoth. 2004, 54, 46–55. [Google Scholar] [CrossRef]
- Han, Q. Control Effect and mechanism of triazole fungicides folicur and caramba on Fusarium head blight and wheat Stripe Rust. Northwest Sci-Tech Univ. Agric. For. 2003, 96, 1712–1728. [Google Scholar]
- Cai, T.T.; Hua, B.X.; Luo, D.W.; Xu, L.R.; Cheng, Q.Y.; Yuan, G.S.; Yan, Z.Q.; Sun, N.; Hua, L.C.; Lu, C. The circadian protein CLOCK regulates cell metabolism via the mitochondrial carrier SLC25A10. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; She, M.Y.; Wang, K.; Riaz, B.; Ye, X.G. Biological roles of ornithine aminotransferase (OAT) in plant stress tolerance: Present progress and future perspectives. Int. J. Mol. Sci. 2018, 19, 3681. [Google Scholar] [CrossRef] [PubMed]
- Swigonova, Z.; Mohsen, A.W.; Vockley, J. Acyl-CoA dehydrogenases: Dynamic history of protein family evolution. J. Mol. Evol. 2009, 69, 176–193. [Google Scholar] [CrossRef] [PubMed]
- Camoes, F.; Islinger, M.; Guimaraes, S.C.; Kilaru, S.; Schuster, M.; Godinho, L.F.; Steinberg, G.; Schrader, M. New insights into the peroxisomal protein inventory: Acyl-CoA oxidases and -dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 111–125. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, E.; Romaris, F.; Lorenzo, S.; Moreno, J.; Bonay, P.; Ubeira, F.M.; Garate, T. A recombinant enolase from Anisakis simplex is differentially recognized in natural human and mouse experimental infections. Med. Microbiol. Immun. 2006, 195, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.X.; Wan, F.; Sun, Z.Z.; Cheng, C.H.; Ling, R.Z.; Fan, L.F.; Wang, A.L. Effect of phosphorus supplementation on cell viability, anti-oxidative capacity and comparative proteomic profiles of puffer fish (Takifugu obscurus) under low temperature stress. Aquaculture 2016, 452, 200–208. [Google Scholar] [CrossRef]
- Ito, S.; Osanai, T. Single amino acid change in 6-phosphogluconate dehydrogenase from Synechocystis conveys higher affinity for NADP(+) and altered mode of inhibition by NADPH. Plant Cell Physiol. 2018, 59, 2452–2461. [Google Scholar] [CrossRef]
- Khan, A.R.; Ullah, I.; Waqas, M.; Shahzad, R.; Hong, S.J.; Park, G.S.; Jung, B.K.; Lee, I.J.; Shin, J.H. Plant growth-promoting potential of endophytic fungi isolated from Solanum nigrum leaves. World J. Microb. Biot. 2015, 31, 1461–1466. [Google Scholar] [CrossRef]
- Delgado, J.; Owens, R.A.; Doyle, S.; Asensio, M.A.; Nunez, F. Impact of the antifungal protein PgAFP from Penicillium chrysogenum on the protein profile in Aspergillus flavus. Appl. Microbiol. Biot. 2015, 99, 8701–8715. [Google Scholar] [CrossRef]
- Lopez-Moya, F.; Suarez-Fernandez, M.; Lopez-Llorca, L.V. Molecular mechanisms of chitosan interactions with fungi and plants. Int. J. Mol. Sci. 2019, 20, 332. [Google Scholar] [CrossRef] [PubMed]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Khullar, S.; Reddy, M.S. Cadmium and arsenic responses in the ectomycorrhizal fungus Laccaria bicolor: Glutathione metabolism and its role in metal(loid) homeostasis. Environ. Microbiol. Rep. 2019, 11, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Lykins, J.D.; Filippova, E.V.; Halavaty, A.S.; Minasov, G.; Zhou, Y.; Dubrovska, I.; Flores, K.J.; Shuvalova, L.A.; Ruan, J.P.; El Bissati, K.; et al. CSGID solves structures and identifies phenotypes for five enzymes in Toxoplasma gondii. Front Cell Infect. Microbiol. 2018, 8, 352. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, M.; Kawasaki, H.; Takenaka, Y.; Kurahashi, O.; Matsui, H. Effects of edd and pgi disruptions on inosine accumulation in Escherichia coli. Biosci. Biotechnol. Biochem. 2005, 69, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Myresiotis, C.K.; Karaoglanidis, G.S.; Tzavella-Monari, K. Resistance of Botrytis cinerea isolates from vegetable crops to anilinopyrimidine, phenylpyrrole, hydroxyanilide, benzimidazole, and dicarboximide fungicides. Plant Dis. 2007, 91, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Walia, S.; Dhingra, S.; Khambay, B.P. Insect growth inhibition, antifeedant and antifungal activity of compounds isolated/derived from Zingiber offcinale Roscoe (ginger) rhizomes. Pest Manag. Sci. 2001, 57, 289–300. [Google Scholar] [CrossRef]
- Xing, C.; Qin, C.Q.; Li, X.Q.; Zhang, F.M.; Linhardt, R.J.; Sun, P.L.; Zhang, A.Q. Chemical composition and biological activities of essential oil isolated by HS-SPME and UAHD from fruits of bergamot. LWT Food Sci. Technol. 2019, 104, 38–44. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–360. [Google Scholar] [CrossRef]
- Luber, C.A.; Cox, J.; Lauterbach, H.; Fancke, B.; Selbach, M.; Tschopp, J.; Akira, S.; Wiegand, M.; Hochrein, H.; O'Keeffe, M.; et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010, 32, 279–289. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.A.; Bertrand, E.M.; Dutkiewicz, S.; Bulygin, V.V.; Moran, D.M.; Monteiro, F.M.; Follows, M.J.; Valois, F.W.; Waterbury, J.B. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. USA 2011, 108, 2184–2189. [Google Scholar] [CrossRef] [PubMed]
- Lange, V.; Malmstrom, J.A.; Didion, J.; King, N.L.; Johansson, B.P.; Schafer, J.; Rameseder, J.; Wong, C.H.; Deutsch, E.W.; Brusniak, M.Y.; et al. Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell. Proteom. 2008, 7, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Gallien, S.; Duriez, E.; Crone, C.; Kellmann, M.; Moehring, T.; Domon, B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell. Proteom. 2012, 11, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]








| Concentration (μg/mL) | Mycelial Diameter (mm) | I (%) |
|---|---|---|
| CK | 33.83 ± 0.54 a | -- |
| 0.0 | 32.67 ± 0.56 a | 3.45 ± 1.65 c |
| 1.0 | 30.83 ± 0.54 a | 8.87 ± 1.60 c |
| 2.0 | 17.33 ± 2.30 b | 48.77 ± 6.81 b |
| 4.0 | 2.33 ± 0.80 c | 93.10 ± 2.37 a |
| 6.0 | 0.83 ± 0.17 c | 97.54 ± 0.49 a |
| Protein | Protein Name | Gene Name | Fold Changes 1 | GO Category 2 |
|---|---|---|---|---|
| A0A0A2KI00 | Uncharacterized protein | PITC_013530 | 9.66 | MF |
| A0A0A2KQ56 | Uncharacterized protein | PITC_017440 | 5.80 | |
| A0A0A2LA04 | Ribulose-phosphate 3-epimerase | PITC_011800 | 3.84 | BP, MF |
| A0A0A2KBU2 | Hydrophobin | PITC_015600 | 3.83 | MF, CC |
| A0A0A2KT64 | Uncharacterized protein | PITC_065480 | 3.15 | |
| A0A0A2KL24 | Uncharacterized protein | PITC_064010 | 2.99 | |
| A0A0A2LAT4 | Uncharacterized protein | PITC_037340 | 2.79 | |
| A0A0A2KR85 | Uncharacterized protein | PITC_014180 | 2.76 | |
| A0A0A2KVZ5 | Uncharacterized protein | PITC_028500 | 2.72 | |
| A0A0A2KMP8 | Uncharacterized protein | PITC_017400 | 2.67 | |
| A0A0A2L522 | Mitochondrial carrier protein | PITC_038210 | 2.51 | MF, CC |
| A0A0A2KX25 | Uncharacterized protein | PITC_051360 | 2.37 | |
| A0A0A2L8T3 | Uncharacterized protein | PITC_098640 | 2.32 | |
| A0A0A2L2N9 | HEAT, type 2 | PITC_085160 | 2.10 | |
| A0A0A2L2V7 | Acyl-CoA dehydrogenase, N-terminal | PITC_066160 | 2.10 | MF |
| A0A0A2LCB9 | Glycoside hydrolase, family 28 | PITC_005000 | 2.10 | BP, MF, CC |
| A0A0A2LAG1 | Xylulose 5-phosphate/Fructose 6-phosphate phosphoketolase, N-terminal | PITC_003850 | 2.10 | BP, MF |
| A0A0A2L8I5 | Glucose-repressible protein Grg1 | PITC_061580 | 2.05 | |
| A0A0A2L934 | Polyketide synthase, enoylreductase | PITC_030400 | 2.04 | MF |
| A0A0A2LBT9 | CDR ABC transporter | PITC_091640 | 0.49 | MF, CC |
| A0A0A2KM09 | ATPase, AAA-type, core | PITC_028850 | 0.48 | MF |
| A0A0A2KZ16 | Mandelate racemase/muconate lactonizing enzyme | PITC_098150 | 0.48 | BP, MF |
| A0A0A2KNF6 | Cytochrome P450 | PITC_065080 | 0.47 | BP, MF |
| A0A0A2LD14 | Short-chain dehydrogenase/reductase SDR | PITC_006840 | 0.46 | |
| A0A0A2LAZ8 | Uncharacterized protein | PITC_023560 | 0.45 | CC |
| A0A0A2KLM8 | Aldo/keto reductase | PITC_048920 | 0.43 | MF |
| A0A0A2L925 | ABC transporter, integral membrane type 1 | PITC_030250 | 0.41 | MF, CC |
| A0A0A2L7S7 | Reverse transcriptase | PITC_011060 | 0.38 | MF |
| A0A0A2L743 | Uncharacterized protein | PITC_032240 | 0.35 | MF |
| A0A0A2KIE8 | Male sterility, NAD-binding | PITC_056110 | 0.34 | MF |
| A0A0A2LCN6 | Short-chain dehydrogenase/reductase SDR | PITC_019130 | 0.31 | |
| A0A0A2LGP0 | Carbon-nitrogen hydrolase | PITC_029870 | 0.30 | BP, MF |
| A0A0A2KZU8 | Peptidase M20 | PITC_086250 | 0.23 | MF |
| A0A0A2LFP8 | O-methyltransferase, family 3 | PITC_025440 | 0.21 | MF |
| A0A0A2L830 | ThiJ/PfpI | PITC_001680 | 0.14 | |
| A0A0A2L1V6 | N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D | PITC_051890 | 0.12 | MF |
| A0A0A2LCF5 | Acyl-transferase/acyl-hydrolase/lysophospholipase | PITC_005350 | 0.08 | MF |
| A0A0A2L5Z5 | Uncharacterized protein | PITC_002480 | 0.08 | |
| A0A0A2L3A1 | Uncharacterized protein | PITC_038310 | 0.07 | |
| A0A0A2LCE5 | Uncharacterized protein | PITC_005250 | 0.06 | |
| A0A0A2LAC2 | Uncharacterized protein | PITC_005360 | 0.04 | |
| A0A0A2LKC5 | Monooxygenase, FAD-binding | PITC_005370 | 0.03 | MF, CC |
| A0A0A2L9W9 | Uncharacterized protein | PITC_005390 | 0.03 | BP, MF |
| A0A0A2KD20 | Uncharacterized protein | PITC_008180 | 0.03 | BP, MF |
| A0A0A2LCH9 | Uncharacterized protein | PITC_005280 | 0.03 | |
| A0A0A2LEG0 | E3 ubiquitin protein ligase | PITC_068020 | + | BP, MF, CC |
| A0A0A2KL74 | Uncharacterized protein | PITC_064410 | + | |
| A0A0A2L3H1 | Kinetochore-Ndc80 subunit Spc24 | PITC_084180 | + | |
| A0A0A2LNS4 | Zinc finger, RING-type | PITC_024830 | + | MF |
| A0A0A2KNZ0 | Translation elongation factor EF1B/ribosomal protein S6 | PITC_014590 | + | MF, CC |
| A0A0A2KWT6 | Ribosome-releasing factor 2, mitochondrial | MEF2 | + | BP, MF, CC |
| A0A0A2LC54 | Peptidase C19, ubiquitin carboxyl-terminal hydrolase 2 | PITC_059690 | + | BP, MF |
| A0A0A2LG71 | Pectin lyase fold/virulence factor | PITC_050450 | + | MF |
| A0A0A2L5T7 | Uncharacterized protein | PITC_001930 | + | BP |
| A0A0A2K9T6 | Uncharacterized conserved protein UCP022603 | PITC_093160 | + | |
| A0A0A2K9G7 | Glycoside hydrolase, superfamily | PITC_049080 | + | BP, MF |
| A0A0A2LF07 | CheY-like superfamily | PITC_061050 | + | MF |
| A0A0A2LN85 | Taxilin family | PITC_010570 | + | MF |
| A0A0A2KY31 | Beta-glucosidase | PITC_028190 | + | BP, MF |
| A0A0A2KE07 | Chromatin-remodeling complex, RSC SWI/SNF subunit Rsc7/Swp82 | PITC_055960 | + | |
| A0A0A2LB02 | RNA polymerase II subunit A | PITC_092370 | + | BP, MF, CC |
| A0A0A2L857 | Transcription factor, MADS-box | PITC_001660 | + | BP, MF, CC |
| A0A0A2L6J4 | Neurolysin/Thimet oligopeptidase, N-terminal | PITC_084000 | + | BP, MF, CC |
| A0A0A2KT14 | Monopolin complex, subunit Csm1/Pcs1 | PITC_066000 | + | |
| A0A0A2LE82 | Uncharacterized protein | PITC_037500 | + | |
| A0A0A2LBD7 | Sof1-like protein | PITC_089150 | + | |
| A0A0A2L5N2 | Striatin, N-terminal | PITC_021600 | + | |
| A0A0A2L4U0 | tRNA-dihydrouridine(47)synthase [NAD(P)(+)] | PITC_047170 | + | MF |
| A0A0A2L4G7 | Phosphatidylserine decarboxylase proenzyme 1, mitochondrial | PSD1 | + | BP, MF, CC |
| A0A0A2KS93 | BolA protein | PITC_042910 | + | |
| A0A0A2LEA3 | Uncharacterized protein | PITC_034190 | + | |
| A0A0A2KWV6 | Sensitivity to Red Light Reduced-like, SRR1 | PITC_077860 | + | |
| A0A0A2L4C5 | Poly(A) polymerase | PITC_097130 | + | BP, MF, CC |
| A0A0A2L1S8 | Heme-containing dehydratase | PITC_040090 | + | |
| A0A0A2L7N4 | Winged helix-turn-helix transcription repressor DNA-binding | PITC_006550 | + | BP, MF, CC |
| A0A0A2KI48 | Aldolase-type TIM barrel | PITC_009330 | + | MF |
| A0A0A2KWA6 | Putative domain, di-copper center | PITC_096580 | + | MF |
| A0A0A2LFQ2 | Uncharacterized protein | PITC_025560 | + | CC |
| A0A0A2LAP4 | Uncharacterized protein | PITC_036940 | + | |
| A0A0A2KHA2 | DNA polymerase III, clamp loader complex, gamma/delta/delta subunit, C-terminal | PITC_071920 | + | BP, MF |
| A0A0A2LFW6 | Catalase, mono-functional, heme-containing | PITC_041140 | + | BP, MF |
| A0A0A2L4V0 | Sas10/Utp3/C1D | PITC_084500 | + | |
| A0A0A2KMY7 | PfkB | PITC_046750 | + | |
| A0A0A2L452 | Blastomyces yeast-phase-specific protein | PITC_083580 | + | |
| A0A0A2LFY2 | Outer membrane protein, IML2, mitochondrial/Tetratricopeptide repeat protein 39 | PITC_041270 | + | CC |
| A0A0A2L4F0 | Vacuolar protein sorting-associated protein 28 | PITC_032200 | + | BP, CC |
| A0A0A2KNJ7 | Ferritin | PITC_095310 | + | BP, MF, CC |
| A0A0A2KCE3 | Uncharacterized protein | PITC_093200 | + | |
| A0A0A2L328 | CheY-like superfamily | PITC_099880 | + | BP, MF |
| A0A0A2KWF5 | Ribosomal protein L53, mitochondrial | PITC_063020 | + | CC |
| A0A0A2LGC1 | Uncharacterized protein | PITC_036010 | + | |
| A0A0A2KAJ2 | Sec1-like protein | PITC_055020 | + | BP, CC |
| A0A0A2KLU4 | H/ACA ribonucleoprotein complex, subunit Nop10 | PITC_015880 | + | BP, MF |
| A0A0A2LB80 | Uncharacterized protein | PITC_088770 | + | MF |
| A0A0A2L283 | Vacuole morphology and inheritance protein 14 | PITC_085800 | + | BP, CC |
| A0A0A2LJG8 | Uncharacterized protein | PITC_091330 | + | |
| A0A0A2LDT3 | Scytalone dehydratase | PITC_055360 | + | BP, MF |
| A0A0A2KPK9 | Alpha/gamma-adaptin-binding protein p34 | PITC_045290 | + | |
| A0A0A2LF46 | Glutamyl-tRNA(Gln) amido-transferase subunit B, mitochondrial | PITC_061230 | + | BP, MF, CC |
| A0A0A2LDQ9 | Uncharacterized protein | PITC_055380 | + | |
| A0A0A2KLL9 | Acyl-transferase/acyl-hydrolase/lysophospholipase | PITC_048820 | + | MF |
| A0A0A2KPR9 | Uncharacterized protein | PITC_054110 | + | CC |
| A0A0A2KYQ5 | Magnesium transporter | PITC_061420 | + | CC |
| A0A0A2KM68 | Uncharacterized protein | PITC_079240 | + | |
| A0A0A2LE28 | Carboxymuconolactone decarboxylase | PITC_044010 | + | MF |
| A0A0A2LBC3 | Uncharacterized protein | PITC_016760 | + | |
| A0A0A2LPW9 | Uncharacterized protein | PITC_059180 | + | |
| A0A0A2KYR5 | Uncharacterized protein | PITC_014450 | + | |
| A0A0A2KPB6 | Exon junction complex, Pym | PITC_045720 | + | BP |
| A0A0A2L3C0 | Uncharacterized protein | PITC_002320 | + | |
| A0A0A2LDR6 | Translation Initiation factor eIF-4e | PITC_055210 | + | MF, CC |
| A0A0A2L3Q9 | Uncharacterized protein | PITC_035540 | + | |
| A0A0A2K7Z3 | RTA-like protein | PITC_013070 | -- | CC |
| A0A0A2K9K0 | Uncharacterized protein | PITC_049430 | -- | CC |
| A0A0A2KC81 | Zinc finger, CCCH-type | PITC_079160 | -- | MF |
| A0A0A2KRX9 | FAD dependent oxidoreductase | PITC_020210 | -- | MF |
| A0A0A2KTM9 | AP-3 complex subunit delta | PITC_064680 | -- | BP, CC |
| A0A0A2KTQ5 | TRAM1-like protein | PITC_000730 | -- | CC |
| A0A0A2KY77 | MAP kinase | PITC_057580 | -- | BP, MF, CC |
| A0A0A2L001 | Uncharacterized protein | PITC_075040 | -- | MF, CC |
| A0A0A2L1F3 | Membrane-associated, eicosanoid/glutathione metabolism protein (MAPEG) | PITC_035700 | -- | CC |
| A0A0A2L3U6 | Uncharacterized protein | PITC_083660 | -- | MF |
| A0A0A2L4J0 | RNA recognition motif domain, eukaryote | PITC_097790 | -- | BP, MF, CC |
| A0A0A2L843 | Serine hydrolase FSH | PITC_001510 | -- | MF |
| A0A0A2L8X0 | Glutamyl-tRNA synthetase, class Ib, archaeal/eukaryotic cytosolic | PITC_073010 | -- | BP, MF, CC |
| A0A0A2L929 | Acyl-CoA dehydrogenase, N-terminal | PITC_030080 | -- | MF |
| A0A0A2L9E5 | Major facilitator superfamily domain, general substrate transporter | PITC_090610 | -- | BP, CC |
| A0A0A2LCH4 | Major facilitator superfamily domain, general substrate transporter | PITC_004620 | -- | MF, CC |
| A0A0A2LEL6 | Tetratricopeptide-like helical | PITC_067660 | -- | MF |
| Comparisons | Number of Involved Proteins | |
|---|---|---|
| Increased | Decreased | |
| Significantly changing in abundance | 19 | 26 |
| Newly arising/undetectable 1 | 67 | 17 |
| Pathway | Protein Name |
|---|---|
| Dioxin degradation | Monooxygenase, FAD-binding |
| Polycyclic aromatic hydrocarbon degradation | Monooxygenase, FAD-binding |
| Linoleic acid metabolism | Cytochrome P450 |
| Betalain biosynthesis | Putative domain, di-copper center |
| Sphingolipid metabolism | TRAM1-like protein |
| Mineral absorption | Ferritin |
| Longevity regulating pathway | Catalase, mono-functional, heme-containing Translation Initiation factor eIF-4e |
| Pentose and glucuronate interconversions | Ribulose-phosphate 3-epimerase Glycoside hydrolase, family 28 |
| MAPK signaling pathway - yeast | Transcription factor, MADS-box; Catalase, mono-functional, heme-containing; MAP kinase |
| mRNA surveillance pathway | RNA polymerase II subunit A; Poly(A) polymerase; Exon junction complex, Pym |
| Carbon fixation in photosynthetic organisms | Ribulose-phosphate 3-epimerase; Xylulose 5-phosphate/Fructose 6-phosphate phosphoketolase, N-terminal |
| Pentose phosphate pathway | Ribulose-phosphate 3-epimerase; Xylulose 5-phosphate/Fructose 6-phosphate phosphoketolase, N-terminal |
| Phenylpropanoid biosynthesis | Beta-glucosidase |
| Naphthalene degradation | Monooxygenase, FAD-binding |
| MAPK signaling pathway - plant | Catalase, mono-functional, heme-containing |
| Chemical carcinogenesis | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Mismatch repair | DNA polymerase III, clamp loader complex, gamma/delta/delta subunit, C-terminal |
| Ferroptosis | Ferritin |
| Isoquinoline alkaloid biosynthesis | Putative domain, di-copper center |
| Cyanoamino acid metabolism | Beta-glucosidase |
| EGFR tyrosine kinase inhibitor resistance | Translation Initiation factor eIF-4e |
| Drug metabolism - cytochrome P450 | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Amyotrophic lateral sclerosis (ALS) | Catalase, mono-functional, heme-containing |
| DNA replication | DNA polymerase III, clamp loader complex, gamma/delta/delta subunit, C-terminal |
| Metabolism of xenobiotics by cytochrome P450 | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Melanogenesis | Putative domain, di-copper centre |
| Platinum drug resistance | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Galactose metabolism | Mandelate racemase/muconate lactonizing enzyme |
| Two-component system | MAP kinase |
| MAPK signaling pathway - fly | Striatin, N-terminal |
| Nucleotide excision repair | DNA polymerase III, clamp loader complex, gamma/delta/delta subunit, C-terminal |
| HIF-1 signaling pathway | Translation Initiation factor eIF-4e |
| Hepatocellular carcinoma | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Lysosome | AP-3 complex subunit delta |
| FoxO signaling pathway | Catalase, mono-functional, heme-containing |
| Fluid shear stress and atherosclerosis | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Longevity regulating pathway - worm | Catalase, mono-functional, heme-containing |
| Drug metabolism - other enzymes | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Necroptosis | Ferritin |
| Longevity regulating pathway–multiple species | Catalase, mono-functional, heme-containing |
| Human T-cell leukemia virus 1 infection | Vacuole morphology and inheritance protein 14 |
| Glutathione metabolism | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Tyrosine metabolism | Putative domain, di-copper center |
| Tryptophan metabolism | Catalase, mono-functional, heme-containing |
| Peroxisome | Catalase, mono-functional, heme-containing |
| Glycerophospholipid metabolism | Phosphatidylserine decarboxylase proenzyme 1, mitochondrial |
| Ribosome biogenesis in eukaryotes | HEAT, type 2,H/ACA ribonucleoprotein complex, subunit Nop10 |
| Insulin signaling pathway | Translation Initiation factor eIF-4e |
| Starch and sucrose metabolism | Beta-glucosidase |
| PI3K-Akt signaling pathway | Translation Initiation factor eIF-4e |
| mTOR signaling pathway | Translation Initiation factor eIF-4e |
| Viral carcinogenesis | Vacuole morphology and inheritance protein 14 |
| Pathways in cancer | Membrane-associated, eicosanoid/glutathione metabolism (MAPEG) protein |
| Amino sugar and nucleotide sugar metabolism | Glycoside hydrolase, superfamily |
| Glyoxylate and dicarboxylate metabolism | Catalase, mono-functional, heme-containing |
| Aminoacyl-tRNA biosynthesis | Glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial |
| RNA transport | Exon junction complex, Pym; Translation Initiation factor eIF-4e |
| Autophagy–yeast | Sec1-like protein |
| Endocytosis | Vacuolar protein sorting-associated protein 28 |
| Spliceosome | Uncharacterized protein |
| Protein Name | Gene Name | Signature Peptides | Proteomics Result 1 | PRM Result 2 |
|---|---|---|---|---|
| Nitro-reductase family protein 3 | PITC_008180 | LGPLLQSHK SISPALWEK | 0.03 | 0.05 |
| Cytochrome P450 | PITC_065080 | AVLSDAVALVR | 0.47 | 0.35 |
| Monooxygenase | PITC_005370 | GALEEALQTYSGVR QLENYDEVDDK SDANTSNAYWNGFHPR | 0.03 | 0.16 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Zhang, X.; Li, M.; Li, T.; Duan, X.; Zhang, D.; Hu, L.; Huang, R. Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum. Int. J. Mol. Sci. 2019, 20, 3459. https://doi.org/10.3390/ijms20143459
Guo M, Zhang X, Li M, Li T, Duan X, Zhang D, Hu L, Huang R. Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum. International Journal of Molecular Sciences. 2019; 20(14):3459. https://doi.org/10.3390/ijms20143459
Chicago/Turabian StyleGuo, Meixia, Xiaoyong Zhang, Meiying Li, Taotao Li, Xuewu Duan, Dandan Zhang, Lianmei Hu, and Riming Huang. 2019. "Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum" International Journal of Molecular Sciences 20, no. 14: 3459. https://doi.org/10.3390/ijms20143459
APA StyleGuo, M., Zhang, X., Li, M., Li, T., Duan, X., Zhang, D., Hu, L., & Huang, R. (2019). Label-Free Proteomic Analysis of Molecular Effects of 2-Methoxy-1,4-naphthoquinone on Penicillium italicum. International Journal of Molecular Sciences, 20(14), 3459. https://doi.org/10.3390/ijms20143459