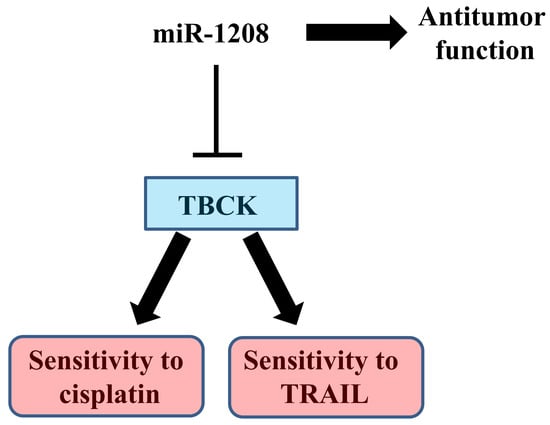
MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. MiR-1208 Inhibits Cell Proliferation and Promotes Apoptosis in Caki-1 Cells
2.2. MiR-1208 Reduces TBCK Expression by Directly Targeting Its 3′-UTR
2.3. MiR-1208 Sensitizes Cisplatin-Induced Apoptosis in Renal Cancer Cells
2.4. MiR-1208 Sensitizes TRAIL-induced Apoptosis in Renal Cancer Cells
2.5. MiR-1208 Plus Cisplatin- and MiR-1208 Plus TRAIL-Induced Apoptosis were Involved in Activation of Caspase-Dependent Apoptotic Pathways
2.6. TBCK Knockdown Improves the Sensitivities to Cisplatin and TRAIL
3. Discussion
4. Materials and Methods
4.1. Cells and Materials
4.2. Western Blot Analysis
4.3. Cell Viability Assay
4.4. Flow Cytometry Analysis
4.5. RNA Isolation and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
4.6. Luciferase Reporter Assays
4.7. Terminal Deoxynucleotidyl Transferase-Mediated dUTP Fluorescein Nick end Labeling (TUNEL) Assay
4.8. Cell Death Assessment by DNA Fragmentation Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer. 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Goga, A.; Benz, C. Anti-oncomir suppression of tumor phenotypes. Mol. Interv. 2007, 7, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meese, E.; Lenhof, H.P.; Keller, A. A dictionary on microRNAs and their putative target pathways. Nucleic. Acids Res. 2010, 38, 4476–4486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Hussein, M.H.; Al-Qahtani, S.A.M.; Shaalan, A.A.M. MicroRNA-34a: A Key Regulator in the Hallmarks of Renal Cell Carcinoma. Oxid. Med. Cell Longev. 2017, 2017, 3269379. [Google Scholar] [CrossRef] [PubMed]
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal cell carcinoma. BMJ 2014, 349, g4797. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Campbell, S.C.; Choi, H.Y.; Jacqmin, D.; Lee, J.E.; Weikert, S.; Kiemeney, L.A. The epidemiology of renal cell carcinoma. Eur. Urol. 2011, 60, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Albiges, L.; Sonpavde, G. Optimal management of metastatic renal cell carcinoma: Current status. Drugs 2013, 73, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, L.; Zhou, H.; Zhao, H.; Zhou, Y.; Xu, R.; Xu, X.; Zheng, L.; Xue, Z.; Xia, W.; Zhang, B.; et al. MicroRNA-99a induces G1-phase cell cycle arrest and suppresses tumorigenicity in renal cell carcinoma. BMC Cancer. 2012, 12, 546. [Google Scholar] [CrossRef]
- Ding, J.; Yeh, C.R.; Sun, Y.; Lin, C.; Chou, J.; Ou, Z.; Chang, C.; Qi, J.; Yeh, S. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 2018, 37, 5037–5053. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.R.; Zhang, Z.Y.; Wang, R.H.; Li, Y.; Mao, Q.Z. MiR-376a functions as tumor suppressor by targeting SGK3 in renal cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3726–3732. [Google Scholar] [PubMed]
- Qin, Z.; Wei, X.; Jin, N.; Wang, Y.; Zhao, R.; Hu, Y.; Yan, W.; Li, J.; Zhou, Q. MiR-199a targeting ROCK1 to affect kidney cell proliferation, invasion and apoptosis. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1920–1925. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhao, L.; Quan, J.; Liu, K.; Lai, Y.; Li, Z.; Zhang, Z.; Xu, J.; Xu, W.; Guan, X.; et al. MiR-378a-5p acts as a tumor suppressor in renal cell carcinoma and is associated with the good prognosis of patients. Am. J. Transl. Res. 2019, 11, 2207–2218. [Google Scholar] [PubMed]
- Xu, X.W.; Li, S.; Yin, F.; Qin, L.L. Expression of miR-205 in renal cell carcinoma and its association with clinicopathological features and prognosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 662–670. [Google Scholar]
- Jiao, D.; Wu, M.; Ji, L.; Liu, F.; Liu, Y. MicroRNA-186 Suppresses Cell Proliferation and Metastasis Through Targeting Sentrin-Specific Protease 1 in Renal Cell Carcinoma. Oncol. Res. 2018, 26, 249–259. [Google Scholar] [CrossRef]
- Kim, E.A.; Kim, S.W.; Nam, J.; Sung, E.G.; Song, I.H.; Kim, J.Y.; Kwon, T.K.; Lee, T.J. Inhibition of c-FLIPL expression by miRNA-708 increases the sensitivity of renal cancer cells to anti-cancer drugs. Oncotarget 2016, 7, 31832–31846. [Google Scholar] [CrossRef]
- Kim, E.A.; Kim, T.G.; Sung, E.G.; Song, I.H.; Kim, J.Y.; Doh, K.O.; Lee, T.J. miR-148a increases the sensitivity to cisplatin by targeting Rab14 in renal cancer cells. Int. J. Oncol. 2017, 50, 984–992. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [Green Version]
- Huppi, K.; Volfovsky, N.; Mackiewicz, M.; Runfola, T.; Jones, T.L.; Martin, S.E.; Stephens, R.; Caplen, N.J. MicroRNAs and genomic instability. Semin Cancer Biol. 2007, 17, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, X.; Liu, W.; Li, B.; Chen, D.; Hu, F.; Wang, L.; Liu, X.M.; Cui, R.; Liu, R. MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene 2019, 39, 4820–4834. [Google Scholar] [CrossRef]
- Huppi, K.; Volfovsky, N.; Runfola, T.; Jones, T.L.; Mackiewicz, M.; Martin, S.E.; Mushinski, J.F.; Stephens, R.; Caplen, N.J. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res. 2008, 6, 212–221. [Google Scholar] [CrossRef]
- Anauate, A.C.; Leal, M.F.; Wisnieski, F.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Calcagno, D.Q.; Assumpção, P.P.; Demachki, S.; Arasaki, C.H.; et al. Analysis of 8q24.21 miRNA cluster expression and copy number variation in gastric cancer. Future Med. Chem. 2019, 11, 947–958. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, X.; Zhou, T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS ONE 2013, 8, e71349. [Google Scholar] [CrossRef]
- Wang, J.; Raimondo, M.; Guha, S.; Chen, J.; Diao, L.; Dong, X.; Wallace, M.B.; Killary, A.M.; Frazier, M.L.; Woodward, T.A.; et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J. Cancer. 2014, 5, 696–705. [Google Scholar] [CrossRef] [Green Version]
- Barsotti, A.M.; Beckerman, R.; Laptenko, O.; Huppi, K.; Caplen, N.J.; Prives, C. p53-Dependent induction of PVT1 and miR-1204. J. Biol Chem. 2012, 287, 2509–2519. [Google Scholar] [CrossRef]
- Shtivelman, E.; Bishop, J.M. The PVT gene frequently amplifies with MYC in tumor cells. Mol. Cell Biol. 1989, 9, 1148–1154. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar, U.; Maeda, H.; Jain, R.K.; Sevick-Muraca, E.M.; Zamboni, W.; Farokhzad, O.C.; Barry, S.T.; Gabizon, A.; Grodzinski, P.; Blakey, D.C. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013, 73, 2412–2417. [Google Scholar] [CrossRef]
- Tivnan, A.; Orr, W.S.; Gubala, V.; Nooney, R.; Williams, D.E.; McDonagh, C.; Prenter, S.; Harvey, H.; Domingo-Fernández, R.; Bray, I.M.; et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS ONE 2012, 7, e38129. [Google Scholar] [CrossRef]
- Shi, L.; Middleton, J.; Jeon, Y.J.; Magee, P.; Veneziano, D.; Laganà, A.; Leong, H.S.; Sahoo, S.; Fassan, M.; Booton, R.; et al. KRAS induces lung tumorigenesis through microRNAs modulation. Cell Death Dis. 2018, 9, 219. [Google Scholar] [CrossRef]
- Gilam, A.; Conde, J.; Weissglas-Volkov, D.; Oliva, N.; Friedman, E.; Artzi, N.; Shomron, N. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat. Commun. 2016, 7, 12868. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.-A.; Jang, J.-H.; Sung, E.-G.; Song, I.-H.; Kim, J.-Y.; Lee, T.-J. MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3540. https://doi.org/10.3390/ijms20143540
Kim E-A, Jang J-H, Sung E-G, Song I-H, Kim J-Y, Lee T-J. MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. International Journal of Molecular Sciences. 2019; 20(14):3540. https://doi.org/10.3390/ijms20143540
Chicago/Turabian StyleKim, Eun-Ae, Ji-Hoon Jang, Eon-Gi Sung, In-Hwan Song, Joo-Young Kim, and Tae-Jin Lee. 2019. "MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells" International Journal of Molecular Sciences 20, no. 14: 3540. https://doi.org/10.3390/ijms20143540
APA StyleKim, E. -A., Jang, J. -H., Sung, E. -G., Song, I. -H., Kim, J. -Y., & Lee, T. -J. (2019). MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. International Journal of Molecular Sciences, 20(14), 3540. https://doi.org/10.3390/ijms20143540