Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1
Abstract
:1. Introduction
2. Claudins and Angulins
2.1. Claudins
2.2. Angulins
3. Claudin and Angulin Binders
3.1. First-Generation Binders: Fragments of Bacterial Toxins
3.2. Second-Generation Binders: Monoclonal Antibodies
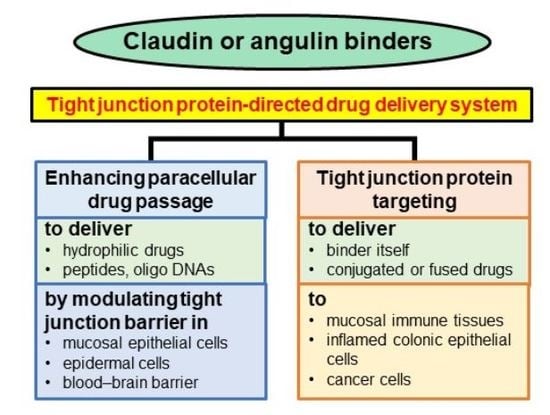
4. Drug Delivery Using Claudin and Angulin Binders
4.1. Mucosal Absorption
4.2. Epidermal Absorption
4.3. Cancer Targeting
4.4. Targeting Tissues Involved in Immunological Processes
4.5. Targeting Inflamed Tissues
4.6. Drug Deliavery to the Brain
5. Safety of Claudin- and Angulin-Targeted Therapies
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | Blood–brain barrier |
| CPE | Clostridium perfringens enterotoxin |
| CPE-R | CPE receptor |
| C-CPE | C-terminal domain of CPE |
| C-CPE-PSIF | C-CPE fused with protein synthesis inhibitory factor |
| ECH | Extracellular helix |
| KD | Knockdown |
| KO | Knockout |
| mAb | Monoclonal antibody |
| MALT | Mucosa-associated lymphoid tissues |
| RVP1 | Rat androgen withdrawal apoptosis protein |
| TER | Transepithelial/transendothelial electrical resistance |
| TJ | Tight junction |
| TM | Transmembrane |
| TNF-α | Tumor necrosis factor-α |
References
- Farquhar, M.G.; Palade, G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963, 17, 375–412. [Google Scholar] [CrossRef] [PubMed]
- de Souza, H.S.P.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastro. Hepat. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Madsen, K.L.; Malfair, D.; Gray, D.; Doyle, J.S.; Jewell, L.D.; Fedorak, R.N. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm. Bowel Dis. 1999, 5, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Tokumasu, R.; Yamaga, K.; Yamazaki, Y.; Murota, H.; Suzuki, K.; Tamura, A.; Bando, K.; Furuta, Y.; Katayama, I.; Tsukita, S. Dose-dependent role of claudin-1 in vivo in orchestrating features of atopic dermatitis. Proc. Natl. Acad. Sci. USA 2016, 113, E4061–E4068. [Google Scholar] [CrossRef] [PubMed]
- Menard, C.; Pfau, M.L.; Hodes, G.E.; Kana, V.; Wang, V.X.; Bouchard, S.; Takahashi, A.; Flanigan, M.E.; Aleyasin, H.; LeClair, K.B.; et al. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci. 2017, 20, 1752–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, C.; Kealy, J.; Humphries, M.M.; Gong, Y.; Hou, J.; Hudson, N.; Cassidy, L.M.; Martiniano, R.; Shashi, V.; Hooper, S.R.; et al. Dose-dependent expression of claudin-5 is a modifying factor in schizophrenia. Mol. Psychiatry 2018, 23, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, L.A.; Mukherjee, T.M.; Williams, A.W. Freeze-etch appearance of tight junctions in epithelium of small and large intestine mice. Protoplasma 1969, 67, 165–184. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Tietgens, A.J.; Anderson, J.M. Visualizing the dynamic coupling of claudin strands to the actin cytoskeleton through ZO-1. Mol. Biol. Cell 2017, 28, 524–534. [Google Scholar] [CrossRef]
- Umeda, K.; Ikenouchi, J.; Katahira-Tayama, S.; Furuse, K.; Sasaki, H.; Nakayama, M.; Matsui, T.; Tsukita, S.; Furuse, M.; Tsukita, S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 2006, 126, 741–754. [Google Scholar] [CrossRef]
- Cording, J.; Berg, J.; Kading, N.; Bellmann, C.; Tscheik, C.; Westphal, J.K.; Milatz, S.; Günzel, D.; Wolburg, H.; Piontek, J.; et al. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J. Cell Sci. 2013, 126, 554–564. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, Y.; Tokumasu, R.; Kimura, H.; Tsukita, S. Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol. Biol. Cell 2011, 22, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Rossa, J.; Ploeger, C.; Vorreiter, F.; Saleh, T.; Protze, J.; Günzel, D.; Wolburg, H.; Krause, G.; Piontek, J. Claudin-3 and claudin-5 protein folding and assembly into the tight junction are controlled by non-conserved residues in the transmembrane 3 (TM3) and extracellular loop 2 (ECL2) segments. J. Biol. Chem. 2014, 289, 7641–7653. [Google Scholar] [CrossRef] [PubMed]
- Tervonen, A.; Ihalainen, T.O.; Nymark, S.; Hyttinen, J. Structural dynamics of tight junctions modulate the properties of the epithelial barrier. PLoS ONE 2019, 14, e0214876. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, L.A. Further observations on fine-structure of freeze-cleaved tight junctions. J. Cell Sci. 1973, 13, 763–786. [Google Scholar] [PubMed]
- Lindmark, T.; Söderholm, J.D.; Olaison, G.; Alvan, G.; Ocklind, G.; Artursson, P. Mechanism of absorption enhancement in humans after rectal administration of ampicillin in suppositories containing sodium caprate. Pharm. Res. 1997, 14, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, N.D.; Miner, M.E.; Hall, W.A.; Siegal, T.; Hanson, E.J.; Osztie, E.; McAllister, L.D.; Bubalo, J.S.; Kraemer, D.F.; Fortin, D.; et al. Safety and efficacy of a multicenter study using intraarterial chemotherapy in conjunction with osmotic opening of the blood-brain barrier for the treatment of patients with malignant brain tumors. Cancer 2000, 88, 637–647. [Google Scholar] [CrossRef]
- Kondoh, M.; Masuyama, A.; Takahashi, A.; Asano, N.; Mizuguchi, H.; Koizumi, N.; Fujii, M.; Hayakawa, T.; Horiguchi, Y.; Watanbe, Y. A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol. Pharmacol. 2005, 67, 749–756. [Google Scholar] [CrossRef]
- Rapoport, S. Osmotic opening of the blood-brain barrier: Principles, mechanism, and therapeutic applications. Cell. Mol. Neurobiol. 2000, 20, 217–230. [Google Scholar] [CrossRef]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Nishizawa, T.; Tani, K.; Yamazaki, Y.; Tamura, A.; Ishitani, R.; Dohmae, N.; Tsukita, S.; Nureki, O.; Fujiyoshi, Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 2014, 344, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, Y.; Suzuki, H.; Tani, K.; Nishikawa, K.; Irie, K.; Ogura, Y.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 2015, 347, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, T.; Shinya, N.; Ito, K.; Ohsawa, N.; Terada, T.; Hirata, K.; Kawano, Y.; Yamamoto, M.; Kimura-Someya, T.; Yokoyama, S.; et al. Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin. Sci. Rep. 2016, 6, 33632. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Irie, K.; Tanaka, H.; Nishikawa, K.; Suzuki, H.; Saitoh, Y.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat. Commun. 2019, 10, 816. [Google Scholar] [CrossRef] [PubMed]
- Piontek, J.; Winkler, L.; Wolburg, H.; Müller, S.L.; Zuleger, N.; Piehl, C.; Wiesner, B.; Krause, G.; Blasig, I.E. Formation of tight junction: Determinants of homophilic interaction between classic claudins. FASEB J. 2008, 22, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tani, K.; Tamura, A.; Tsukita, S.; Fujiyoshi, Y. Model for the architecture of claudin-based paracellular ion channels through tight junctions. J. Mol. Biol. 2015, 427, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Ikenouchi, J.; Furuse, M.; Furuse, K.; Sasaki, H.; Tsukita, S.; Tsukita, S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005, 171, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Oda, Y.; Sasaki, H.; Ikenouchi, J.; Higashi, T.; Akashi, M.; Nishi, E.; Furuse, M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J. Cell Sci. 2011, 124, 548–555. [Google Scholar] [CrossRef] [Green Version]
- Higashi, T.; Tokuda, S.; Kitajiri, S.; Masuda, S.; Nakamura, H.; Oda, Y.; Furuse, M. Analysis of the ‘angulin’ proteins LSR, ILDR1 and ILDR2-tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J. Cell Sci. 2013, 126, 966–977. [Google Scholar] [CrossRef]
- Nakatsu, D.; Kano, F.; Taguchi, Y.; Sugawara, T.; Nishizono, T.; Nishikawa, K.; Oda, Y.; Furuse, M.; Murata, M. JNK1/2-dependent phosphorylation of angulin-1/LSR is required for the exclusive localization of angulin-1/LSR and tricellulin at tricellular contacts in EpH4 epithelial sheet. Genes Cells 2014, 19, 565–581. [Google Scholar] [CrossRef]
- Kim, N.K.D.; Higashi, T.; Lee, K.Y.; Kim, A.R.; Kitajiri, S.; Kim, M.Y.; Chang, M.Y.; Kim, V.; Oh, S.H.; Kim, D.; et al. Downsloping high-frequency hearing loss due to inner ear tricellular tight junction disruption by a novel ILDR1 mutation in the Ig-like domain. PLoS ONE 2015, 10, e0116931. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Yagi, K.; Kondoh, M. Current progress in a second-generation claudin binder, anti-claudin antibody, for clinical applications. Drug Discov. Today 2016, 21, 1711–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, Y.; Yagi, K.; Kondoh, M. Roles of the first-generation claudin binder, Clostridium perfringens enterotoxin, in the diagnosis and claudin-targeted treatment of epithelium-derived cancers. Pflugers Arch. 2017, 469, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Okada, Y.; Shirakura, K.; Tachibana, K.; Sawada, M.; Yagi, K.; Doi, T.; Kondoh, M. Anti-claudin antibodies as a concept for development of claudin-directed drugs. J. Pharmacol. Exp. Ther. 2019, 368, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Kitadokoro, K.; Nishimura, K.; Kamitani, S.; Fukui-Miyazaki, A.; Toshima, H.; Abe, H.; Kamata, Y.; Sugita-Konishi, Y.; Yamamoto, S.; Karatani, H.; et al. Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J. Biol. Chem. 2011, 286, 19549–19555. [Google Scholar] [CrossRef] [PubMed]
- Katahira, J.; Inoue, N.; Horiguchi, Y.; Matsuda, M.; Sugimoto, N. Molecular cloning and functional characterization of the receptor for Clostridium perfringensenterotoxin. J. Cell Biol. 1997, 136, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Furuse, M.; Fujimoto, K.; Tsukita, S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. USA 1999, 96, 511–516. [Google Scholar] [CrossRef] [Green Version]
- Sonoda, N.; Furuse, M.; Sasaki, H.; Yonemura, S.; Katahira, J.; Horiguchi, Y.; Tsukita, S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 1999, 147, 195–204. [Google Scholar] [CrossRef]
- Winkler, L.; Gehring, C.; Wenzel, A.; Müller, S.L.; Piehl, C.; Krause, G.; Blasig, I.E.; Piontek, J. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 2009, 284, 18863–18872. [Google Scholar] [CrossRef]
- Fujita, K.; Katahira, J.; Horiguchi, Y.; Sonoda, N.; Furuse, M.; Tsukita, S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000, 476, 258–261. [Google Scholar] [CrossRef]
- Uchida, H.; Kondoh, M.; Hanada, T.; Takahashi, A.; Hamakubo, T.; Yagi, K. A claudin-4 modulator enhances the mucosal absorption of a biologically active peptide. Biochem. Pharmacol. 2010, 79, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Komiya, E.; Kakutani, H.; Yoshida, T.; Fujii, M.; Horiguchi, Y.; Mizuquchi, H.; Tsutsumi, Y.; Tsunoda, S.I.; Koizumi, N.; et al. Domain mapping of a claudin-4 modulator, the C-terminal region of C-terminal fragment of Clostridium perfringens enterotoxin, by site-directed mutagenesis. Biochem. Pharmacol. 2008, 75, 1639–1648. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Saito, Y.; Kondoh, M.; Matsushita, K.; Krug, S.M.; Suzuki, H.; Tsujino, H.; Li, X.R.; Aoyama, H.; Matsuhisa, K.; et al. Creation and biochemical analysis of a broad-specific claudin binder. Biomaterials 2012, 33, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Veshnyakova, A.; Piontek, J.; Protze, J.; Waziri, N.; Heise, I.; Krause, G. Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J. Biol. Chem. 2012, 287, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Protze, J.; Eichner, M.; Piontek, A.; Dinter, S.; Rossa, J.; Blecharz, K.G.Z.; Vajkoczy, P.; Piontek, J.; Krause, G. Directed structural modification of Clostridium perfringens enterotoxin to enhance binding to claudin-5. Cell. Mol. Life Sci. 2015, 72, 1417–1432. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, W.; Piontek, A.; Protze, J.; Eichner, M.; Mahringer, A.; Subileau, E.A.; Lee, I.F.M.; Schulzke, J.D.; Krause, G.; Piontek, J. Reversible opening of the blood-brain barrier by claudin-5-binding variants of Clostridium perfringens enterotoxin’s claudin-binding domain. Biomaterials 2018, 161, 129–143. [Google Scholar] [CrossRef]
- Nagahama, M.; Umezaki, M.; Oda, M.; Kobayashi, K.; Tone, S.; Suda, T.; Ishidoh, K.; Sakurai, J. Clostridium perfringens iota-toxin b induces rapid cell necrosis. Infect. Immun. 2011, 79, 4353–4360. [Google Scholar] [CrossRef]
- Nagahama, M.; Yamaguchi, A.; Hagiyama, T.; Ohkubo, N.; Kobayashi, K.; Sakurai, J. Binding and internalization of Clostridium perfringens iota-toxin in lipid rafts. Infect. Immun. 2004, 72, 3267–3275. [Google Scholar] [CrossRef]
- Papatheodorou, P.; Carette, J.E.; Bell, G.W.; Schwan, C.; Guttenberg, G.; Brummelkamp, T.R.; Aktories, K. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc. Natl. Acad. Sci. USA 2011, 108, 16422–16427. [Google Scholar] [CrossRef]
- Krug, S.M.; Hayaishi, T.; Iguchi, D.; Watari, A.; Takahashi, A.; Fromm, M.; Nagahama, M.; Takeda, H.; Okada, Y.; Sawasaki, T.; et al. Angubindin-1, a novel paracellular absorption enhancer acting at the tricellular tight junction. J. Control. Release 2017, 260, 1–11. [Google Scholar] [CrossRef]
- Souriau, C.; Hudson, P.J. Recombinant antibodies for cancer diagnosis and therapy. Expert Opin. Biol. Ther. 2003, 3, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Espiritu, M.J.; Collier, A.C.; Bingham, J.P. A 21st-century approach to age-old problems: The ascension of biologics in clinical therapeutics. Drug Discov. Today 2014, 19, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Nagase, S.; Iida, M.; Takeda, S.; Yamashita, M.; Watari, A.; Shirasago, Y.; Fukasawa, M.; Takeda, H.; Sawasaki, T.; et al. Claudin-1 binder enhances epidermal permeability in a human keratinocyte model. J. Pharmacol. Exp. Ther. 2015, 354, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Shirakura, K.; Okada, Y.; Takeda, H.; Endo, K.; Tamura, M.; Watari, A.; Sadamura, Y.; Sawasaki, T.; Doi, T.; et al. Claudin-5-binders enhance permeation of solutes across the blood-brain barrier in a mammalian model. J. Pharmacol. Exp. Ther. 2017, 363, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Zhou, W.; Hamauchi, K.; Shirakura, K.; Doi, T.; Yagi, K.; Sawasaki, T.; Okada, Y.; Kondoh, M.; Takeda, H. Engineered membrane protein antigens successfully induce antibodies against extracellular regions of claudin-5. Sci. Rep. 2018, 8, 8383. [Google Scholar] [CrossRef]
- Takigawa, M.; Iida, M.; Nagase, S.; Suzuki, H.; Watari, A.; Tada, M.; Okada, Y.; Doi, T.; Fukasawa, M.; Yagi, K.; et al. Creation of a claudin-2 binder and its tight junction-modulating activity in a human intestinal models. J. Pharmacol. Exp. Ther. 2017, 363, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Fofana, I.; Krieger, S.E.; Grunert, F.; Glauben, S.; Xiao, F.; Fafi-Kremer, S.; Soulier, E.; Royer, C.; Thumann, C.; Mee, C.J.; et al. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 2010, 139, 953–964. [Google Scholar] [CrossRef]
- Fukasawa, M.; Nagase, S.; Shirasago, Y.; Iida, M.; Yamashita, M.; Endo, K.; Yagi, K.; Suzuki, T.; Wakita, T.; Hanada, K.; et al. Monoclonal antibodies against extracellular domains of claudin-1 block hepatitis C virus infection in a mouse model. J. Virol. 2015, 89, 4866–4879. [Google Scholar] [CrossRef]
- Shimizu, Y.; Shirasago, Y.; Kondoh, M.; Suzuki, T.; Wakita, T.; Hanada, K.; Yagi, K.; Fukasawa, M. Monoclonal antibodies against occludin completely prevented hepatitis C virus infection in a mouse model. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Okai, K.; Ichikawa-Tomikawa, N.; Saito, A.C.; Watabe, T.; Sugimoto, K.; Fujita, D.; Ono, C.; Fukuhara, T.; Matsuura, Y.; Ohira, H.; et al. A novel occludin-targeting monoclonal antibody prevents hepatitis C virus infection in vitro. Oncotarget 2018, 9, 16588–16598. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; Tada, M.; Iida, M.; Nagase, S.; Hata, T.; Watari, A.; Okada, Y.; Doi, T.; Fukasawa, M.; Yagi, K.; et al. Generation and characterization of a human-mouse chimeric antibody against the extracellular domain of claudin-1 for cancer therapy using a mouse model. Biochem. Biophys. Res. Commun. 2016, 477, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Cherradi, S.; Ayrolles-Torro, A.; Vezzo-Vie, N.; Gueguinou, N.; Denis, V.; Combes, E.; Boissiere, F.; Busson, M.; Canterel-Thouennon, L.; Mollevi, C.; et al. Antibody targeting of claudin-1 as a potential colorectal cancer therapy. J. Exp. Clin. Cancer Res. 2017, 36, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, Y.; Hata, T.; Tada, M.; Iida, M.; Watari, A.; Okada, Y.; Doi, T.; Kuniyasu, H.; Yagi, K.; Kondoh, M. Safety evaluation of a human chimeric monoclonal antibody that recognizes the extracellular loop domain of claudin-2. Eur. J. Pharm. Sci. 2018, 117, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Suzuki, M.; Kato-Nakano, M.; Kawamoto, S.; Misaka, H.; Kimoto, N.; Furuya, A.; Nakamura, K. Generation of specific monoclonal antibodies against the extracellular loops of human claudin-3 by immunizing mice with target-expressing cells. Biosci. Biotechnol. Biochem. 2015, 79, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Romani, C.; Cocco, E.; Bignotti, E.; Moratto, D.; Bugatti, A.; Todeschini, P.; Bandiera, E.; Tassi, R.; Zanotti, L.; Pecorelli, S.; et al. Evaluation of a novel human IgG1 anti-claudin3 antibody that specifically recognizes its aberrantly localized antigen in ovarian cancer cells and that is suitable for selective drug delivery. Oncotarget 2015, 6, 34617–34628. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Kato-Nakano, M.; Kawamoto, S.; Furuya, A.; Abe, Y.; Misaka, H.; Kimoto, N.; Nakamura, K.; Ohta, S.; Ando, H. Therapeutic antitumor efficacy of monoclonal antibody against Claudin-4 for pancreatic and ovarian cancers. Cancer Sci. 2009, 100, 1623–1630. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Kawahigashi, Y.; Hata, T.; Li, X.; Watari, A.; Tada, M.; Ishii-Watabe, A.; Okada, Y.; Doi, T.; Fukasawa, M.; et al. Efficacy and safety evaluation of claudin-4-targeted antitumor therapy using a human and mouse cross-reactive monoclonal antibody. Pharmacol. Res. Perspect. 2016, 4, e00266. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Serada, S.; Enomoto, T.; Takahashi, Y.; Nakagawa, S.; Nojima, S.; Morimoto, A.; Matsuzaki, S.; Yokoyama, T.; Takahashi, T.; et al. LSR antibody therapy inhibits ovarian epithelial tumor growth by inhibiting lipid uptake. Cancer Res. 2018, 78, 516–527. [Google Scholar] [CrossRef]
- Ben-David, U.; Nudel, N.; Benvenisty, N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat. Commun. 2013, 4, 1992. [Google Scholar] [CrossRef]
- Powell, D.W. Barrier Function of Epithelia. Am. J. Physiol. 1981, 241, G275–G288. [Google Scholar] [CrossRef]
- Tran, T.N. Cutaneous drug delivery: An update. J. Investig. Dermatol. Symp. Proc. 2013, 16, S67–S69. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Furuse, M. Claudin-based barrier in simple and stratified cellular sheets. Curr. Opin. Cell Biol. 2002, 14, 531–536. [Google Scholar] [CrossRef]
- Furuse, M.; Hata, M.; Furuse, K.; Yoshida, Y.; Haratake, A.; Sugitani, Y.; Noda, T.; Kubo, A.; Tsukita, S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: A lesson from claudin-1-deficient mice. J. Cell Biol. 2002, 156, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Osanai, M.; Takasawa, A.; Murata, M.; Sawada, N. Claudins in cancer: Bench to bedside. Pflugers Arch. 2017, 469, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.S.; Morgado-Diaz, J.A. Claudins: Multifunctional players in epithelial tight junctions and their role in cancer. Cell. Mol. Life Sci. 2007, 64, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Long, H.Y.; Crean, C.D.; Lee, W.H.; Cummings, O.W.; Gabig, T.G. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001, 61, 7878–7881. [Google Scholar] [PubMed]
- Kominsky, S.L.; Vali, M.; Korz, D.; Gabig, T.G.; Weitzman, S.A.; Argani, P.; Sukumar, S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol. 2004, 164, 1627–1633. [Google Scholar] [CrossRef]
- Michl, P.; Barth, C.; Buchholz, M.; Lerch, M.M.; Rolke, M.; Holzmann, K.H.; Menke, A.; Fensterer, H.; Giehl, K.; Lohr, M.; et al. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003, 63, 6265–6271. [Google Scholar]
- Zhu, Y.H.; Brannstrom, M.; Janson, P.O.; Sundfeldt, K. Differences in expression patterns of the tight junction proteins, claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int. J. Cancer 2006, 118, 1884–1891. [Google Scholar] [CrossRef]
- Weber, C.R.; Nalle, S.C.; Tretiakova, M.; Rubin, D.T.; Turner, J.R. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab. Investig. 2008, 88, 1110–1120. [Google Scholar] [CrossRef] [Green Version]
- Kinugasa, T.; Huo, Q.; Higash, D.; Shibaguchi, H.; Kuroki, M.; Tanaka, T.; Futami, K.; Yamashita, Y.; Hachimine, K.; Maekawa, S.; et al. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res. 2007, 27, 3729–3734. [Google Scholar] [CrossRef]
- Dhawan, P.; Singh, A.B.; Deane, N.G.; No, Y.; Shiou, S.R.; Schmidt, C.; Neff, J.; Washington, M.K.; Beauchamp, R.D. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Investig. 2005, 115, 1765–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhawan, P.; Ahmad, R.; Chaturvedi, R.; Smith, J.J.; Midha, R.; Mittal, M.K.; Krishnan, M.; Chen, X.; Eschrich, S.; Yeatman, T.J.; et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: Role of epidermal growth factor receptor activation. Oncogene 2011, 30, 3234–3247. [Google Scholar] [CrossRef] [PubMed]
- Tabaries, S.; Dong, Z.; Annis, M.G.; Omeroglu, A.; Pepin, F.; Ouellet, V.; Russo, C.; Hassanain, M.; Metrakos, P.; Diaz, Z.; et al. Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 2011, 30, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Fukasawa, M.; Kuniyasu, H.; Yagi, K.; Kondoh, M. Claudin-targeted drug development using anti-claudin monoclonal antibodies to treat hepatitis and cancer. Ann. N. Y. Acad. Sci. 2017, 1397, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Michl, P.; Buchholz, M.; Rolke, M.; Kunsch, S.; Lohr, M.; McClane, B.; Tsukita, S.; Leder, G.; Adler, G.; Gress, T.M. Claudin-4: A new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001, 121, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J. Claudin proteins in human cancer: Promising new targets for diagnosis and therapy. Cancer Res. 2005, 65, 9603–9606. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, S.L. Claudins: Emerging targets for cancer therapy. Expert Rev. Mol. Med. 2006, 8, 1–11. [Google Scholar] [CrossRef]
- Saeki, R.; Kondoh, M.; Kakutani, H.; Tsunoda, S.; Mochizuki, Y.; Hamakubo, T.; Tsutsumi, Y.; Horiguchi, Y.; Yagi, K. A novel tumor-targeted therapy using a claudin-4-targeting molecule. Mol. Pharmacol. 2009, 76, 918–926. [Google Scholar] [CrossRef]
- Meunier, V.; Bourrie, M.; Berger, Y.; Fabre, G. The human intestinal epithelial cell line Caco-2; pharmacological and pharmacokinetic applications. Cell Biol. Toxicol. 1995, 11, 187–194. [Google Scholar] [CrossRef]
- Torres, J.B.; Knight, J.C.; Mosley, M.J.; Kersemans, V.; Koustoulidou, S.; Allen, D.; Kinchesh, P.; Smart, S.; Cornelissen, B. Imaging of claudin-4 in pancreatic ductal adenocarcinoma using a radiolabelled anti-claudin-4 monoclonal antibody. Mol. Imaging Biol. 2018, 20, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Kunisawa, J.; Fukuyama, S.; Kiyono, H. Mucosa-associated lymphoid tissues in the aerodigestive tract: Their shared and divergent traits and their importance to the orchestration of the mucosal immune system. Curr. Mol. Med. 2005, 5, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Kraehenbuhl, J.P.; Neutra, M.R. Epithelial M cells: Differentiation and function. Annu. Rev. Cell Dev. Biol. 2000, 16, 301–332. [Google Scholar] [CrossRef] [PubMed]
- Nagatake, T.; Fujita, H.; Minato, N.; Hamazaki, Y. Enteroendocrine cells are specifically marked by cell surface expression of claudin-4 in mouse small intestine. PLoS ONE 2014, 9, e90638. [Google Scholar] [CrossRef] [PubMed]
- Tamagawa, H.; Takahashi, I.; Furuse, M.; Yoshitake-Kitano, Y.; Tsukita, S.; Ito, T.; Matsuda, H.; Kiyono, H. Characteristics of claudin expression in follicle-associated epithelium of Peyer’s patches: Preferential localization of claudin-4 at the apex of the dome region. Lab. Investig. 2003, 83, 1045–1053. [Google Scholar] [CrossRef]
- Ye, T.; Yue, Y.; Fan, X.M.; Dong, C.S.; Xu, W.; Xiong, S.D. M cell-targeting strategy facilitates mucosal immune response and enhances protection against CVB3-induced viral myocarditis elicited by chitosan-DNA vaccine. Vaccine 2014, 32, 4457–4465. [Google Scholar] [CrossRef]
- Kakutani, H.; Kondoh, M.; Fukasaka, M.; Suzuki, H.; Hamakubo, T.; Yagi, K. Mucosal vaccination using claudin-4-targeting. Biomaterials 2010, 31, 5463–5471. [Google Scholar] [CrossRef]
- Suzuki, H.; Kakutani, H.; Kondoh, M.; Watari, A.; Yagi, K. The safety of a mucosal vaccine using the C-terminal fragment of Clostridium perfringens enterotoxin. Pharmazie 2010, 65, 766–769. [Google Scholar] [CrossRef]
- Suzuki, H.; Hosomi, K.; Nasu, A.; Kondoh, M.; Kunisawa, J. Development of adjuvant-free bivalent food poisoning vaccine by augmenting the antigenicity of Clostridium perfringens enterotoxin. Front. Immunol. 2018, 9, 2320. [Google Scholar] [CrossRef]
- Mankertz, J.; Amasheh, M.; Krug, S.M.; Fromm, A.; Amasheh, S.; Hillenbrand, B.; Tavalali, S.; Fromm, M.; Schulzke, J.D. TNF alpha up-regulates claudin-2 expression in epithelial HT-29/B6 cells via phosphatidylinositol-3-kinase signaling. Cell Tissue Res. 2009, 336, 67–77. [Google Scholar] [CrossRef]
- Furuse, M.; Furuse, K.; Sasaki, H.; Tsukita, S. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J. Cell Biol. 2001, 153, 263–272. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood-brain barrier: Bottleneck in brain drug development. NeuroRx 2005, 2, 3–14. [Google Scholar] [CrossRef]
- Vanlandewijck, M.; He, L.; Mae, M.A.; Andrae, J.; Ando, K.; Del Gaudio, F.; Nahar, K.; Lebouvier, T.; Lavina, B.; Gouveia, L.; et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018, 554, 475–480. [Google Scholar] [CrossRef] [Green Version]
- Nitta, T.; Hata, M.; Gotoh, S.; Seo, Y.; Sasaki, H.; Hashimoto, N.; Furuse, M.; Tsukita, S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J. Cell Biol. 2003, 161, 653–660. [Google Scholar] [CrossRef]
- Sohet, F.; Lin, C.; Munji, R.N.; Lee, S.Y.; Ruderisch, N.; Soung, A.; Arnold, T.D.; Derugin, N.; Vexler, Z.S.; Yen, F.T.; et al. LSR/angulin-1 is a tricellular tight junction protein involved in blood-brain barrier formation. J. Cell Biol. 2015, 208, 703–711. [Google Scholar] [CrossRef]
- Zeniya, S.; Kuwahara, H.; Daizo, K.; Watari, A.; Kondoh, M.; Yoshida-Tanaka, K.; Kaburagi, H.; Asada, K.; Nagata, T.; Nagahama, M.; et al. Angubindin-1 opens the blood-brain barrier in vivo for delivery of antisense oligonucleotide to the central nervous system. J. Control. Release 2018, 283, 126–134. [Google Scholar] [CrossRef]
- Fujita, H.; Hamazaki, Y.; Noda, Y.; Oshima, M.; Minato, N. Claudin-4 deficiency results in urothelial hyperplasia and lethal hydronephrosis. PLoS ONE 2012, 7, e52272. [Google Scholar] [CrossRef]
- Muto, S.; Hata, M.; Taniguchi, J.; Tsuruoka, S.; Moriwaki, K.; Saitou, M.; Furuse, K.; Sasaki, H.; Fujimura, A.; Imai, M.; et al. Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal proximal tubules. Proc. Natl. Acad. Sci. USA 2010, 107, 8011–8016. [Google Scholar] [CrossRef]
- Gong, Y.F.; Himmerkus, N.; Sunq, A.; Milatz, S.; Merkel, C.; Bleich, M.; Hou, J.H. ILDR1 is important for paracellular water transport and urine concentration mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, 5271–5276. [Google Scholar] [CrossRef] [Green Version]
- Hadj-Rabia, S.; Baala, L.; Vabres, P.; Hamel-Teillac, D.; Jacquemin, E.; Fabre, M.; Lyonnet, S.; De Prost, Y.; Munnich, A.; Hadchouel, M.; et al. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with lchthyosis: A tight junction disease. Gastroenterology 2004, 127, 1386–1390. [Google Scholar] [CrossRef]
- Arinami, T. Analyses of the associations between the genes of 22q11 deletion syndrome and schizophrenia. J. Hum. Genet. 2006, 51, 1037–1045. [Google Scholar] [CrossRef]
- Wei, J.; Hemmings, G.P. A study of the combined effect of the CLDN5 locus and the genes for the phospholipid metabolism pathway in schizophrenia. Prostaglandins Leukot. Essent. Fatty Acids 2005, 73, 441–445. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Wei, J.; Xie, L.; Shen, Y.; Liu, S.Z.; Ju, G.Z.; Shi, J.P.; Yu, Y.Q.; Zhang, X.; Xu, Q.; et al. The CLDN5 locus may be involved in the vulnerability to schizophrenia. Eur. Psychiatry 2004, 19, 354–357. [Google Scholar] [CrossRef]
- Tanaka, H.; Imasato, M.; Yamazaki, Y.; Matsumoto, K.; Kunimoto, K.; Delpierre, J.; Meyer, K.; Zerial, M.; Kitamura, N.; Watanabe, M.; et al. Claudin-3 regulates bile canalicular paracellular barrier and cholesterol gallstone cores formation in mice. J. Hepatol. 2018, 69, 1308–1316. [Google Scholar] [CrossRef]
- Furuse, M.; Sasaki, H.; Tsukita, S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999, 147, 891–903. [Google Scholar] [CrossRef]
- Knipp, G.T.; Ho, N.F.H.; Barsuhn, C.L.; Borchardt, R.T. Paracellular diffusion in Caco-2 cell monolayers: Effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. J. Pharm. Sci. 1997, 86, 1105–1110. [Google Scholar] [CrossRef]
- Watson, C.J.; Rowland, M.; Warhurst, G. Functional modeling of tight junctions in intestinal cell monolayers using polyethylene glycol oligomers. Am. J. Physiol. Cell Physiol. 2001, 281, C388–C397. [Google Scholar] [CrossRef]
- Steinke, A.; Meier-Stiegen, S.; Drenckhahn, D.; Asan, E. Molecular composition of tight and adherens junctions in the rat olfactory epithelium and fila. Histochem. Cell Biol. 2008, 130, 339–361. [Google Scholar] [CrossRef]
- Schlingmann, B.; Overgaard, C.E.; Molina, S.A.; Lynn, K.S.; Mitchell, L.A.; Dorsainvil White, S.; Mattheyses, A.L.; Guidot, D.M.; Capaldo, C.T.; Koval, M. Regulation of claudin/zonula occludens-1 complexes by hetero-claudin interactions. Nat. Commun. 2016, 7, 12276. [Google Scholar] [CrossRef] [Green Version]
- Markov, A.G.; Veshnyakova, A.; Fromm, M.; Amasheh, M.; Amasheh, S. Segmental expression of claudin proteins correlates with tight junction barrier properties in rat intestine. J. Comp. Physiol. B 2010, 180, 591–598. [Google Scholar] [CrossRef]
- Inai, T.; Sengoku, A.; Guan, X.; Hirose, E.; Iida, H.; Shibata, Y. Heterogeneity in expression and subcellular localization of tight junction proteins, claudin-10 and -15, examined by RT-PCR and immunofluorescence microscopy. Arch. Histol. Cytol. 2005, 68, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Rahner, C.; Mitic, L.L.; Anderson, J.M. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology 2001, 120, 411–422. [Google Scholar] [CrossRef]
- Kiuchi-Saishin, Y.; Gotoh, S.; Furuse, M.; Takasuga, A.; Tano, Y.; Tsukita, S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J. Am. Soc. Nephrol. 2002, 13, 875–886. [Google Scholar]
- Steinemann, A.; Galm, I.; Chip, S.; Nitsch, C.; Maly, L.P. Claudin-1, -2 and -3 are selectively expressed in the epithelia of the choroid plexus of the mouse from early development and into adulthood while claudin-5 is restricted to endothelial cells. Front. Neuroanat. 2016, 10, 16. [Google Scholar] [CrossRef]
- He, L.Q.; Vanlandewijck, M.; Mae, M.A.; Andrae, J.; Ando, K.; Del Gaudio, F.; Nahar, K.; Lebouvier, T.; Lavina, B.; Gouveia, L.; et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 2018, 5, 180160. [Google Scholar] [CrossRef] [Green Version]
- Watari, A.; Kodaka, M.; Matsuhisa, K.; Sakamoto, Y.; Hisaie, K.; Kawashita, N.; Takagi, T.; Yamagishi, Y.; Suzuki, H.; Tsujino, H.; et al. Identification of claudin-4 binder that attenuates tight junction barrier function by TR-FRET-based screening assay. Sci. Rep. 2017, 7, 14514. [Google Scholar] [CrossRef] [Green Version]





| Binder Type | Mutated Regions | Ref. |
|---|---|---|
| Negative-binding mutant | Y306A/L315A | [42] |
| Broad-spectrum binder (m19) | S304A/S305P/S307R/N309H/S313H | [43] |
| Enhancing specificity to claudin-3 | L223A/D225A/R227A | [44] |
| Enhancing specificity to claudin-4 | L254A/S256A/I258A/D284A | [44] |
| Enhancing specificity to claudin-5 | Y306W/S313H | [45] |
| Improved specificity to claudin-5 | N218Q/Y306W/S313H | [46] |
| Indicated Application or Disease | Target | Monoclonal Antibody Name | Ref. or ClinicalTrials.gov Identifier |
|---|---|---|---|
| Modulation of epidermal barrier | Claudin-1 | 7A5 | [53] |
| Modulation of blood–brain barrier | Claudin-5 | R9, R2, 2B12 | [54,55] |
| Inflammatory bowel disease | Claudin-2 | 1A2 | [56] |
| Hepatitis C virus infection | Claudin-1 | OM-7D3-B3, 3A2 | [57,58] |
| Occludin | 1-3, 67-2 | [59,60] | |
| Gastric cancer (phase III study) | Claudin-18.2 | IMAB362 | NCT03504397 |
| Pancreatic cancer (phase II study) | Claudin-18.2 | IMAB362 | NCT03816163 |
| Germ cell tumor (phase II study) | Claudin-6 | IMAB027 | NCT03760081 |
| Cancers (phase I or pre-clinical study) | Claudin-1 | 3A2, 6F6 | [61,62] |
| Claudin-2 | 1A2 | [63] | |
| Claudin-3 | KMK3953, IgGH6 | [64,65] | |
| Claudin-4 | KM3934, 5D12 | [66,67] | |
| Angulin-1 | #1-25 | [68] | |
| Angulin-3 | BAY1905254 | NCT03666273 | |
| Regenerative medicine | Claudin-6 | clone 342927 | [69] |
| Phenotype of Knockout (KO) or Knockdown (KD) Mice | Ref. | |
|---|---|---|
| Claudin-1 | Atopic dermatitis (KD) | [4] |
| Claudin-2 | Impaired renal Na+, Cl−, and water reabsorption (KO) | [108] |
| Claudin-3 | Increased hepatocyte permeability to phosphate ion (KO) | [114] |
| Claudin-4 | Impaired renal Ca2+ and Cl− reabsorption (KO) | [107] |
| Claudin-5 | Schizophrenia-like symptoms (KD) | [6] |
| Angulin-2 | Impaired renal water reabsorption and colonic water absorption (KO) | [109] |
| Claudin | Angulin | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | |||
| Epidermal cells (stratum granulosum) | + | - | - | + | - | + | - | - | [29,73] | |
| Nasal epithelial cells | + | - | + | + | + | + | + | - | [29,118] | |
| Lung (alveoli) | + | - | + | + | + | + | - | - | [29,119] | |
| Small intestine (jejunum) | + | + | + | - | + | + | - | - | [29,109,120] | |
| Colon (surface) | + | - | + | + | + | - | + | - | [29,109,120] | |
| Liver | + | + | + | - | - | + | - | - | [29,121,122] | |
| Kidney (glomerulus) | + | + | - | - | - | - | + | - | [29,123] | |
| Kidney (proximal tube) | - | + | - | - | - | + | - | - | [29,123] | |
| Kidney (thin ascending limb of the loop of Henle) | - | - | + | + | - | - | + | - | [29,123] | |
| Kidney (collecting duct) | - | - | + | + | - | - | + | - | [29,109,123] | |
| Brain endothelial cells | - | - | - | - | + | + | - | + | [103,105] | |
| Brain ependymal cells | + | + | + | - | - | - | - | + | [29,124] | |
| Lung endothelial cells | - | - | - | - | + | - | - | - | [125] | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto, Y.; Tachibana, K.; Krug, S.M.; Kunisawa, J.; Fromm, M.; Kondoh, M. Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1. Int. J. Mol. Sci. 2019, 20, 4016. https://doi.org/10.3390/ijms20164016
Hashimoto Y, Tachibana K, Krug SM, Kunisawa J, Fromm M, Kondoh M. Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1. International Journal of Molecular Sciences. 2019; 20(16):4016. https://doi.org/10.3390/ijms20164016
Chicago/Turabian StyleHashimoto, Yosuke, Keisuke Tachibana, Susanne M. Krug, Jun Kunisawa, Michael Fromm, and Masuo Kondoh. 2019. "Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1" International Journal of Molecular Sciences 20, no. 16: 4016. https://doi.org/10.3390/ijms20164016
APA StyleHashimoto, Y., Tachibana, K., Krug, S. M., Kunisawa, J., Fromm, M., & Kondoh, M. (2019). Potential for Tight Junction Protein–Directed Drug Development Using Claudin Binders and Angubindin-1. International Journal of Molecular Sciences, 20(16), 4016. https://doi.org/10.3390/ijms20164016