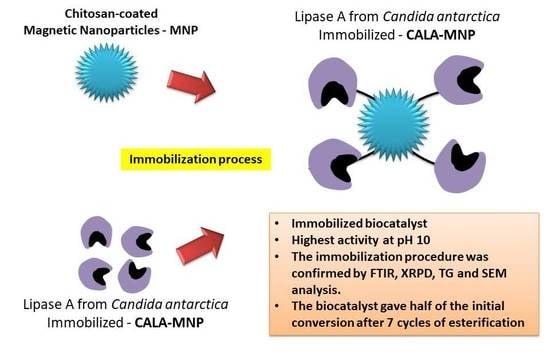
Immobilization of Lipase A from Candida antarctica onto Chitosan-Coated Magnetic Nanoparticles
Abstract
:1. Introduction
2. Results and Discussion
2.1. Immobilization Parameters
2.2. Effect of pH on the Thermal Stability of CALA Biocatalysts
2.3. Effect of pH on CALA Biocatalysts Activity
2.4. Characterization of the Nanoparticles and Biocatalysts
2.5. Operational Stability
3. Materials and Methods
3.1. Materials
3.2. Methods
3.2.1. Synthesis of Iron Magnetic Nanoparticles (Fe3O4) Functionalized with Chitosan (CHI)
3.2.2. Activation of Fe3O4@CHI with Glutaraldehyde (GLU)
3.2.3. Covalent Immobilization of CALA on Fe3O4@CHI-GLU
3.2.4. Immobilization of CALA on Fe3O4@CHI
3.2.5. Determination of Enzymatic Activity and Protein Concentration
3.2.6. Immobilization Parameters
3.2.7. Thermal and pH Inactivation
3.2.8. Effect of pH on Biocatalyst Activity
3.2.9. X-ray Powder Diffraction (XRPD)
3.2.10. Fourier Transform Infrared (FTIR) Spectroscopy
3.2.11. Thermogravimetry (TG)
3.2.12. Scanning Electron Microscope (SEM)
3.2.13. Extraction and Purification of Tilapia Oil
3.2.14. Production of Free Fatty Acids (FFAs) from tilapia oil
3.2.15. Production of Biolubricant Ester
3.2.16. Operational Stability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chowdhury, A.; Mitra, D.; Biswas, D. Biolubricant synthesis from waste cooking oil via enzymatic hydrolysis followed by chemical esterification: Biolubricant synthesis from waste cooking oil. J. Chem. Technol. Biotechnol. 2013, 88, 139–144. [Google Scholar] [CrossRef]
- Linko, Y.-Y.; Lämsä, M.; Huhtala, A.; Linko, P. Lipase-catalyzed transesterification of rapeseed oil and 2-ethyl-1-hexanol. J. Am. Oil Chem. Soc. 1994, 71, 1411–1414. [Google Scholar] [CrossRef]
- Kleinaitė, E.; Jaška, V.; Tvaska, B.; Matijošytė, I. A cleaner approach for biolubricant production using biodiesel as a starting material. J. Clean. Prod. 2014, 75, 40–44. [Google Scholar] [CrossRef]
- Chowdhury, A.; Chakraborty, R.; Mitra, D.; Biswas, D. Optimization of the production parameters of octyl ester biolubricant using Taguchi’s design method and physico-chemical characterization of the product. Ind. Crops Prod. 2014, 52, 783–789. [Google Scholar] [CrossRef]
- Cavalcanti, E.D.C.; Aguieiras, É.C.G.; da Silva, P.R.; Duarte, J.G.; Cipolatti, E.P.; Fernandez-Lafuente, R.; da Silva, J.A.C.; Freire, D.M.G. Improved production of biolubricants from soybean oil and different polyols via esterification reaction catalyzed by immobilized lipase from Candida rugosa. Fuel 2018, 215, 705–713. [Google Scholar] [CrossRef]
- Sharma, B.K.; Biresaw, G. Environmentally Friendly and Biobased Lubricants; CRC Press: Boca Raton, FL, USA, 2017; p. 434. [Google Scholar]
- Fernandes, K.V.; Papadaki, A.; da Silva, J.A.C.; Fernandez-Lafuente, R.; Koutinas, A.A.; Freire, D.M.G. Enzymatic esterification of palm fatty-acid distillate for the production of polyol esters with biolubricant properties. Ind. Crops Prod. 2018, 116, 90–96. [Google Scholar] [CrossRef]
- Da Silva, J.A.C.; Soares, V.F.; Fernandez-Lafuente, R.; Habert, A.C.; Freire, D.M.G. Enzymatic production and characterization of potential biolubricants from castor bean biodiesel. J. Mol. Catal. B Enzym. 2015, 122, 323–329. [Google Scholar] [CrossRef]
- Greco-Duarte, J.; Cavalcanti-Oliveira, E.D.; Da Silva, J.A.C.; Fernandez-Lafuente, R.; Freire, D.M.G. Two-step enzymatic production of environmentally friendly biolubricants using castor oil: Enzyme selection and product characterization. Fuel 2017, 202, 196–205. [Google Scholar] [CrossRef]
- De Oliveira, U.M.F.; de Lima Matos, L.J.B.; de Souza, M.C.M.; Pinheiro, B.B.; dos Santos, J.C.S.; Gonçalves, L.R.B. Effect of the presence of surfactants and immobilization conditions on catalysts’ properties of Rhizomucor miehei Lipase onto Chitosan. Appl. Biochem. Biotechnol. 2018, 184, 1263–1285. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, Y.; Hou, M.; Li, T.; Ge, J.; Liu, Z. Chemo-enzymatic synthesis of valrubicin using Pluronic conjugated lipase with temperature responsiveness in organic media. RSC Adv. 2013, 3, 22963. [Google Scholar] [CrossRef]
- Gharat, N.; Rathod, V.K. Enzyme catalyzed transesterification of waste cooking oil with dimethyl carbonate. J. Mol. Catal. B Enzym. 2013, 88, 36–40. [Google Scholar] [CrossRef]
- Villalba, M.; Verdasco-Martín, C.M.; dos Santos, J.C.S.; Fernandez-Lafuente, R.; Otero, C. Operational stabilities of different chemical derivatives of Novozym 435 in an alcoholysis reaction. Enzyme Microb. Technol. 2016, 90, 35–44. [Google Scholar] [CrossRef]
- Palla, C.A.; Pacheco, C.; Carrín, M.E. Production of structured lipids by acidolysis with immobilized Rhizomucor miehei lipases: Selection of suitable reaction conditions. J. Mol. Catal. B Enzym. 2012, 76, 106–115. [Google Scholar] [CrossRef]
- Törnvall, U.; Orellana-Coca, C.; Hatti-Kaul, R.; Adlercreutz, D. Stability of immobilized Candida antarctica lipase B during chemo-enzymatic epoxidation of fatty acids. Enzyme Microb. Technol. 2007, 40, 447–451. [Google Scholar] [CrossRef]
- Akanbi, T.O.; Barrow, C.J. Candida antarctica lipase A effectively concentrates DHA from fish and thraustochytrid oils. Food Chem. 2017, 229, 509–516. [Google Scholar] [CrossRef]
- Kasche, V.; Haufler, U.; Riechmann, L. Equilibrium and kinetically controlled synthesis with enzymes: Semisynthesis of penicillins and peptides. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1987; pp. 280–292. [Google Scholar]
- Bolsoni-Lopes, A.; Alonso-Vale, M.I.C. Lipolysis and lipases in white adipose tissue—An update. Arch. Endocrinol. Metab. 2015, 59, 335–342. [Google Scholar] [CrossRef]
- Robles-Medina, A.; González-Moreno, P.A.; Esteban-Cerdán, L.; Molina-Grima, E. Biocatalysis: Towards ever greener biodiesel production. Biotechnol. Adv. 2009, 27, 398–408. [Google Scholar] [CrossRef]
- Uppenberg, J.; Patkar, S.; Bergfors, T.; Jones, T.A. Crystallization and preliminary X-ray studies of lipase B from Candida antarctica. J. Mol. Biol. 1994, 235, 790–792. [Google Scholar] [CrossRef]
- Ericsson, D.J.; Kasrayan, A.; Johansson, P.; Bergfors, T.; Sandström, A.G.; Bäckvall, J.-E.; Mowbray, S.L. X-ray Structure of Candida antarctica lipase A shows a novel lid structure and a likely mode of interfacial activation. J. Mol. Biol. 2008, 376, 109–119. [Google Scholar] [CrossRef]
- Zamost, B.L.; Nielsen, H.K.; Starnes, R.L. Thermostable enzymes for industrial applications. J. Ind. Microbiol. 1991, 8, 71–81. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Lokha, Y.; Fernández-Lafuente, R. Immobilization on octyl-agarose beads and some catalytic features of commercial preparations of lipase a from Candida antarctica (Novocor ADL): Comparison with immobilized lipase B from Candida antarctica. Biotechnol. Prog. 2019, 35, e2735. [Google Scholar] [CrossRef]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Bonazza, H.L.; de Matos, L.J.B.L.; Carneiro, E.A.; Barbosa, O.; Fernandez-Lafuente, R.; Gonçalves, L.R.B.; de Sant’ Ana, H.B.; Santiago-Aguiar, R.S. Immobilization of CALB on activated chitosan: Application to enzymatic synthesis in supercritical and near-critical carbon dioxide. Biotechnol. Rep. 2017, 14, 16–26. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, Á.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Hanefeld, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Zainal, N.A.; Zulkifli, N.W.M.; Gulzar, M.; Masjuki, H.H. A review on the chemistry, production, and technological potential of bio-based lubricants. Renew. Sustain. Energy Rev. 2018, 82, 80–102. [Google Scholar] [CrossRef]
- Shahabuddin, M.; Masjuki, H.H.; Kalam, M.A.; Bhuiya, M.M.K.; Mehat, H. Comparative tribological investigation of bio-lubricant formulated from a non-edible oil source (Jatropha oil). Ind. Crops Prod. 2013, 47, 323–330. [Google Scholar] [CrossRef]
- Singh, Y.; Farooq, A.; Raza, A.; Mahmood, M.A.; Jain, S. Sustainability of a non-edible vegetable oil based bio-lubricant for automotive applications: A review. Process Saf. Environ. Prot. 2017, 111, 701–713. [Google Scholar] [CrossRef]
- Munoz, R.A.; Fernandes, D.M.; Santos, D.Q.; Barbosa, T.G.; Sousa, R.M. Biodiesel: Production, characterization, metallic corrosion and analytical methods for contaminants. In Biodiesel—Feedstocks, Production and Applications; InTech Open: London, UK, 2012; pp. 129–175. [Google Scholar]
- Nagendramma, P.; Kaul, S. Development of ecofriendly/biodegradable lubricants: An overview. Renew. Sustain. Energy Rev. 2012, 16, 764–774. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, S. Potential non-edible oil resources as biodiesel feedstock: An Indian perspective. Renew. Sustain. Energy Rev. 2011, 15, 1791–1800. [Google Scholar] [CrossRef]
- Singh, D.; Singh, S.P. Low cost production of ester from non edible oil of Argemone mexicana. Biomass Bioenergy 2010, 34, 545–549. [Google Scholar] [CrossRef]
- Le Clef, E.; Kemper, T. Sunflower seed preparation and oil extraction. In Sunflower; Elsevier: Amsterdam, The Netherlands, 2015; pp. 187–226. [Google Scholar]
- Hamid, H.A.; Yunus, R.; Rashid, U.; Choong, T.S.Y.; Al-Muhtaseb, A.H. Synthesis of palm oil-based trimethylolpropane ester as potential biolubricant: Chemical kinetics modeling. Chem. Eng. J. 2012, 200–202, 532–540. [Google Scholar] [CrossRef]
- Jayadas, N.H.; Nair, K.P.; Ajithkumar, G. Tribological evaluation of coconut oil as an environment-friendly lubricant. Tribol. Int. 2007, 40, 350–354. [Google Scholar] [CrossRef]
- Ting, C.-C.; Chen, C.-C. Viscosity and working efficiency analysis of soybean oil based bio-lubricants. Measurement 2011, 44, 1337–1341. [Google Scholar] [CrossRef]
- Do Valle, C.P.; Rodrigues, J.S.; Fechine, L.M.U.D.; Cunha, A.P.; Queiroz Malveira, J.; Luna, F.M.T.; Ricardo, N.M.P.S. Chemical modification of Tilapia oil for biolubricant applications. J. Clean. Prod. 2018, 191, 158–166. [Google Scholar] [CrossRef]
- Martins, G.I.; Secco, D.; Tokura, L.K.; Bariccatti, R.A.; Dolci, B.D.; Santos, R.F. Potential of tilapia oil and waste in biodiesel production. Renew. Sustain. Energy Rev. 2015, 42, 234–239. [Google Scholar] [CrossRef]
- Roriz, G.D.; Delphino, M.K.D.V.C.; Gardner, I.A.; Gonçalves, V.S.P. Characterization of tilapia farming in net cages at a tropical reservoir in Brazil. Aquac. Rep. 2017, 6, 43–48. [Google Scholar] [CrossRef]
- Silva, J.F.X.; Ribeiro, K.; Silva, J.F.; Cahú, T.B.; Bezerra, R.S. Utilization of tilapia processing waste for the production of fish protein hydrolysate. Animal Feed Sci. Technol. 2014, 196, 96–106. [Google Scholar] [CrossRef]
- Villamil, O.; Váquiro, H.; Solanilla, J.F. Fish viscera protein hydrolysates: Production, potential applications and functional and bioactive properties. Food Chem. 2017, 224, 160–171. [Google Scholar] [CrossRef]
- De Souza, M.C.M.; dos Santos, K.P.; Freire, R.M.; Barreto, A.C.H.; Fechine, P.B.A.; Gonçalves, L.R.B. Production of flavor esters catalyzed by lipase B from Candida antarctica immobilized on magnetic nanoparticles. Braz. J. Chem. Eng. 2017, 34, 681–690. [Google Scholar] [CrossRef]
- Netto, C.G.C.M.; Toma, H.E.; Andrade, L.H. Superparamagnetic nanoparticles as versatile carriers and supporting materials for enzymes. J. Mol. Catal. B Enzym. 2013, 85–86, 71–92. [Google Scholar] [CrossRef]
- Zhou, G.; Fung, K.K.; Wong, L.W.; Chen, Y.; Renneberg, R.; Yang, S. Immobilization of glucose oxidase on rod-like and vesicle-like mesoporous silica for enhancing current responses of glucose biosensors. Talanta 2011, 84, 659–665. [Google Scholar] [CrossRef]
- Li, X.; Zhu, H.; Feng, J.; Zhang, J.; Deng, X.; Zhou, B.; Zhang, H.; Xue, D.; Li, F.; Mellors, N.J.; et al. One-pot polylol synthesis of graphene decorated with size- and density-tunable Fe3O4 nanoparticles for porcine pancreatic lipase immobilization. Carbon 2013, 60, 488–497. [Google Scholar] [CrossRef]
- Cipolatti, E.P.; Valério, A.; Henriques, R.O.; Moritz, D.E.; Ninow, J.L.; Freire, D.M.G.; Manoel, E.A.; Fernandez-Lafuente, R.; de Oliveira, D. Nanomaterials for biocatalyst immobilization—State of the art and future trends. RSC Adv. 2016, 6, 104675–104692. [Google Scholar] [CrossRef]
- Xu, W.; Ling, P.; Zhang, T. Polymeric micelles, a promising drug delivery system to enhance bioavailability of poorly water-soluble drugs. J. Drug Deliv. 2013, 2013, 340315. [Google Scholar] [CrossRef]
- Bilal, M.; Zhao, Y.; Rasheed, T.; Iqbal, H.M.N. Magnetic nanoparticles as versatile carriers for enzymes immobilization: A review. Int. J. Biol. Macromol. 2018, 120, 2530–2544. [Google Scholar] [CrossRef]
- Sinha, V.R.; Singla, A.K.; Wadhawan, S.; Kaushik, R.; Kumria, R.; Bansal, K.; Dhawan, S. Chitosan microspheres as a potential carrier for drugs. Int. J. Pharm. 2004, 274, 1–33. [Google Scholar] [CrossRef]
- Silva, J.A.; Macedo, G.P.; Rodrigues, D.S.; Giordano, R.L.C.; Gonçalves, L.R.B. Immobilization of Candida antarctica lipase B by covalent attachment on chitosan-based hydrogels using different support activation strategies. Biochem. Eng. J. 2012, 60, 16–24. [Google Scholar] [CrossRef]
- Anitha, A.; Sowmya, S.; Kumar, P.T.S.; Deepthi, S.; Chennazhi, K.P.; Ehrlich, H.; Tsurkan, M.; Jayakumar, R. Chitin and chitosan in selected biomedical applications. Prog. Polym. Sci. 2014, 39, 1644–1667. [Google Scholar] [CrossRef]
- Khwaldia, K.; Basta, A.H.; Aloui, H.; El-Saied, H. Chitosan–caseinate bilayer coatings for paper packaging materials. Carbohydr. Polym. 2014, 99, 508–516. [Google Scholar] [CrossRef]
- Badawi, M.A.; Negm, N.A.; Abou Kana, M.T.H.; Hefni, H.H.; Abdel Moneem, M.M. Adsorption of aluminum and lead from wastewater by chitosan-tannic acid modified biopolymers: Isotherms, kinetics, thermodynamics and process mechanism. Int. J. Biol. Macromol. 2017, 99, 465–476. [Google Scholar] [CrossRef]
- Oliveira, H.C.; Gomes, B.C.R.; Pelegrino, M.T.; Seabra, A.B. Nitric oxide-releasing chitosan nanoparticles alleviate the effects of salt stress in maize plants. Nitric Oxide 2016, 61, 10–19. [Google Scholar] [CrossRef]
- Krajewska, B. Application of chitin- and chitosan-based materials for enzyme immobilizations: A review. Enzyme Microb. Technol. 2004, 35, 126–139. [Google Scholar] [CrossRef]
- Pinheiro, B.B.; Rios, N.S.; Rodríguez Aguado, E.; Fernandez-Lafuente, R.; Freire, T.M.; Fechine, P.B.A.; dos Santos, J.C.S.; Gonçalves, L.R.B. Chitosan activated with divinyl sulfone: A new heterofunctional support for enzyme immobilization. Application in the immobilization of lipase B from Candida antarctica. Int. J. Biol. Macromol. 2019, 130, 798–809. [Google Scholar] [CrossRef]
- Nicolás, P.; Saleta, M.; Troiani, H.; Zysler, R.; Lassalle, V.; Ferreira, M.L. Preparation of iron oxide nanoparticles stabilized with biomolecules: Experimental and mechanistic issues. Acta Biomater. 2013, 9, 4754–4762. [Google Scholar] [CrossRef]
- Freire, T.M.; Dutra, L.M.U.; Queiroz, D.C.; Ricardo, N.M.P.S.; Barreto, K.; Denardin, J.C.; Wurm, F.R.; Sousa, C.P.; Correia, A.N.; de Lima-Neto, P.; et al. Fast ultrasound assisted synthesis of chitosan-based magnetite nanocomposites as a modified electrode sensor. Carbohydr. Polym. 2016, 151, 760–769. [Google Scholar] [CrossRef]
- Shete, P.B.; Patil, R.M.; Thorat, N.D.; Prasad, A.; Ningthoujam, R.S.; Ghosh, S.J.; Pawar, S.H. Magnetic chitosan nanocomposite for hyperthermia therapy application: Preparation, characterization and in vitro experiments. Appl. Surf. Sci. 2014, 288, 149–157. [Google Scholar] [CrossRef]
- De Andrades, D.; Graebin, N.G.; Kadowaki, M.K.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Immobilization and stabilization of different β-glucosidases using the glutaraldehyde chemistry: Optimal protocol depends on the enzyme. Int. J. Biol. Macromol. 2019, 129, 672–678. [Google Scholar] [CrossRef]
- Vazquez-Ortega, P.G.; Alcaraz-Fructuoso, M.T.; Rojas-Contreras, J.A.; López-Miranda, J.; Fernandez-Lafuente, R. Stabilization of dimeric β-glucosidase from Aspergillu s nige r via glutaraldehyde immobilization under different conditions. Enzyme Microb. Technol. 2018, 110, 38–45. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Rueda, N.; Barbosa, O.; Fernández-Sánchez, J.F.; Medina-Castillo, A.L.; Ramón-Márquez, T.; Arias-Martos, M.C.; Millán-Linares, M.C.; Pedroche, J.; del Yust, M.M.; et al. Characterization of supports activated with divinyl sulfone as a tool to immobilize and stabilize enzymes via multipoint covalent attachment. Application to chymotrypsin. RSC Adv. 2015, 5, 20639–20649. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Ait Braham, S.; Hussain, F.; Morellon-Sterling, R.; Kamal, S.; Kornecki, J.F.; Barbosa, O.; Kati, D.E.; Fernandez-Lafuente, R. Cooperativity of covalent attachment and ion exchange on alcalase immobilization using glutaraldehyde chemistry: Enzyme stabilization and improved proteolytic activity. Biotechnol. Prog. 2019, 35, e2768. [Google Scholar] [CrossRef]
- Zaak, H.; Peirce, S.; de Albuquerque, T.; Sassi, M.; Fernandez-Lafuente, R. Exploiting the versatility of aminated supports activated with glutaraldehyde to immobilize β-galactosidase from Aspergillus oryzae. Catalysts 2017, 7, 250. [Google Scholar] [CrossRef]
- Osuna, Y.; Sandoval, J.; Saade, H.; López, R.G.; Martinez, J.L.; Colunga, E.M.; de la Cruz, G.; Segura, E.P.; Arévalo, F.J.; Zon, M.A.; et al. Immobilization of Aspergillus niger lipase on chitosan-coated magnetic nanoparticles using two covalent-binding methods. Bioprocess Biosyst. Eng. 2015, 38, 1437–1445. [Google Scholar] [CrossRef]
- Rodrigues, D.S.; Mendes, A.A.; Adriano, W.S.; Gonçalves, L.R.B.; Giordano, R.L. Multipoint covalent immobilization of microbial lipase on chitosan and agarose activated by different methods. J. Mol. Catal. B Enzym. 2008, 51, 100–109. [Google Scholar] [CrossRef]
- Berger, J.; Reist, M.; Mayer, J.M.; Felt, O.; Peppas, N.A.; Gurny, R. Structure and interactions in covalently and ionically crosslinked chitosan hydrogels for biomedical applications. Eur. J. Pharm. Biopharm. 2004, 57, 19–34. [Google Scholar] [CrossRef]
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique Biocatalysts from a Unique Origin. Org. Process Res. Dev. 2002, 6, 446–451. [Google Scholar] [CrossRef]
- Heldt-Hansen, H.P.; Ishii, M.; Patkar, S.A.; Hansen, T.T.; Eigtved, P. A new immobilized positional nonspecific lipase for fat modification and ester synthesis. In Biocatalysis in Agricultural Biotechnology; Whitaker, J.R., Sonnet, P.E., Eds.; American Chemical Society: Washington, DC, USA, 1989; Volume 389, pp. 158–172. ISBN 978-0-8412-1571-9. [Google Scholar]
- Zaak, H.; Fernandez-Lopez, L.; Velasco-Lozano, S.; Alcaraz-Fructuoso, M.T.; Sassi, M.; Lopez-Gallego, F.; Fernandez-Lafuente, R. Effect of high salt concentrations on the stability of immobilized lipases: Dramatic deleterious effects of phosphate anions. Process Biochem. 2017, 62, 128–134. [Google Scholar] [CrossRef]
- Martinelle, M.; Holmquist, M.; Hult, K. On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim. Biophys. Acta Lipids Lipid Metab. 1995, 1258, 272–276. [Google Scholar] [CrossRef]
- Melo, A.; Silva, F.; dos Santos, J.; Fernández-Lafuente, R.; Lemos, T.; Dias Filho, F. Synthesis of benzyl acetate catalyzed by lipase immobilized in nontoxic chitosan-polyphosphate beads. Molecules 2017, 22, 2165. [Google Scholar] [CrossRef]
- Yang, X.; Liu, L.; Zhang, M.; Tan, W.; Qiu, G.; Zheng, L. Improved removal capacity of magnetite for Cr(VI) by electrochemical reduction. J. Hazard. Mater. 2019, 374, 26–34. [Google Scholar] [CrossRef]
- Long, J.; Yu, X.; Xu, E.; Wu, Z.; Xu, X.; Jin, Z.; Jiao, A. In situ synthesis of new magnetite chitosan/carrageenan nanocomposites by electrostatic interactions for protein delivery applications. Carbohydr. Polym. 2015, 131, 98–107. [Google Scholar] [CrossRef]
- El-Guendouz, S.; Aazza, S.; Lyoussi, B.; Bankova, V.; Lourenço, J.; Costa, A.; Mariano, J.; Miguel, M.; Faleiro, M. Impact of biohybrid magnetite nanoparticles and moroccan propolis on adherence of methicillin resistant strains of staphylococcus aureus. Molecules 2016, 21, 1208. [Google Scholar] [CrossRef]
- Rasoulzadeh, H.; Mohseni-Bandpei, A.; Hosseini, M.; Safari, M. Mechanistic investigation of ciprofloxacin recovery by magnetite–imprinted chitosan nanocomposite: Isotherm, kinetic, thermodynamic and reusability studies. Int. J. Biol. Macromol. 2019, 133, 712–721. [Google Scholar] [CrossRef]
- Neto, C.G.T.; Giacometti, J.A.; Job, A.E.; Ferreira, F.C.; Fonseca, J.L.C.; Pereira, M.R. Thermal analysis of chitosan based networks. Carbohydr. Polym. 2005, 62, 97–103. [Google Scholar] [CrossRef]
- Ziegler-Borowska, M.; Chełminiak, D.; Kaczmarek, H. Thermal stability of magnetic nanoparticles coated by blends of modified chitosan and poly(quaternary ammonium) salt. J. Therm. Anal. Calorim. 2015, 119, 499–506. [Google Scholar] [CrossRef]
- Poon, L.; Wilson, L.D.; Headley, J.V. Chitosan-glutaraldehyde copolymers and their sorption properties. Carbohydr. Polym. 2014, 109, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.C.A.; Miranda, J.S.; Bolina, I.C.A.; Silva, W.C.; Hirata, D.B.; de Castro, H.F.; Mendes, A.A. Immobilization of porcine pancreatic lipase on poly-hydroxybutyrate particles for the production of ethyl esters from macaw palm oils and pineapple flavor. Biochem. Eng. J. 2014, 82, 139–149. [Google Scholar] [CrossRef]
- Reis, C.; Sousa, E.; Serpa, J.; Oliveira, R.; Oliveira, R.; Santos, J. Design of immobilized enzyme biocatalysts: Drawbacks and opportunities. Quim. Nova 2019, 42, 768–783. [Google Scholar] [CrossRef]
- Xie, W.; Ma, N. Immobilized lipase on Fe3O4 nanoparticles as biocatalyst for biodiesel production. Energy Fuels 2009, 23, 1347–1353. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Neto, D.M.A.; Galvão, W.S.; Rios, N.S.; Carvalho, A.C.L.D.M.; Correa, M.A.; Bohn, F.; Fernandez-Lafuente, R.; Fechine, P.B.A.; de Mattos, M.C.; et al. Design of a lipase-nano particle biocatalysts and its use in the kinetic resolution of medicament precursors. Biochem. Eng. J. 2017, 125, 104–115. [Google Scholar] [CrossRef]
- De Souza, T.C.; Fonseca, T.D.S.; da Costa, J.A.; Rocha, M.V.P.; de Mattos, M.C.; Fernandez-Lafuente, R.; Gonçalves, L.R.B.; dos Santos, J.C. Cashew apple bagasse as a support for the immobilization of lipase B from Candida antarctica: Application to the chemoenzymatic production of (R)-Indanol. J. Mol. Catal. B Enzym. 2016, 130, 58–69. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ishak, A.A.; Salimon, J. Synthesis of rubber seed oil and trimethylolpropane based biolubricant base stocks. Malays. J. Anal. Sci. 2013, 17, 414–421. [Google Scholar]




| Biocatalyst | IY(%) | AtT (U/g) | AtD (U/g) | AtR (%) |
|---|---|---|---|---|
| Fe3O4@CHI–GLU–CALA | 84.1 ± 1.0 | 212.2 ± 1.0 | 208.0 ± 3.0 | 98.0 ± 3.0 |
| Fe3O4@CHI-CALA | 44.3 ± 1.5 | 123.1 ± 1.5 | 120.8 ± 2.0 | 98.1 ± 2.0 |
| Biocatalyst | Half-Life (t1/2, min) | ||
|---|---|---|---|
| pH 5 | pH 7 | pH 9 | |
| CALA | 10.1 | 5.7 | 27 |
| CALA-MNP | 92 | 62 | 222 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, R.R.C.; Lima, P.J.M.; Pinheiro, B.B.; Freire, T.M.; Dutra, L.M.U.; Fechine, P.B.A.; Gonçalves, L.R.B.; de Souza, M.C.M.; dos Santos, J.C.S.; Fernandez-Lafuente, R. Immobilization of Lipase A from Candida antarctica onto Chitosan-Coated Magnetic Nanoparticles. Int. J. Mol. Sci. 2019, 20, 4018. https://doi.org/10.3390/ijms20164018
Monteiro RRC, Lima PJM, Pinheiro BB, Freire TM, Dutra LMU, Fechine PBA, Gonçalves LRB, de Souza MCM, dos Santos JCS, Fernandez-Lafuente R. Immobilization of Lipase A from Candida antarctica onto Chitosan-Coated Magnetic Nanoparticles. International Journal of Molecular Sciences. 2019; 20(16):4018. https://doi.org/10.3390/ijms20164018
Chicago/Turabian StyleMonteiro, Rodolpho R. C., Paula J. M. Lima, Bruna B. Pinheiro, Tiago M. Freire, Lillian M. U. Dutra, Pierre B. A. Fechine, Luciana R. B. Gonçalves, Maria C. M. de Souza, José C. S. dos Santos, and Roberto Fernandez-Lafuente. 2019. "Immobilization of Lipase A from Candida antarctica onto Chitosan-Coated Magnetic Nanoparticles" International Journal of Molecular Sciences 20, no. 16: 4018. https://doi.org/10.3390/ijms20164018
APA StyleMonteiro, R. R. C., Lima, P. J. M., Pinheiro, B. B., Freire, T. M., Dutra, L. M. U., Fechine, P. B. A., Gonçalves, L. R. B., de Souza, M. C. M., dos Santos, J. C. S., & Fernandez-Lafuente, R. (2019). Immobilization of Lipase A from Candida antarctica onto Chitosan-Coated Magnetic Nanoparticles. International Journal of Molecular Sciences, 20(16), 4018. https://doi.org/10.3390/ijms20164018