Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.)
Abstract
:1. Introduction
2. Results
2.1. Summary of the AhGRF Gene Family in Peanut
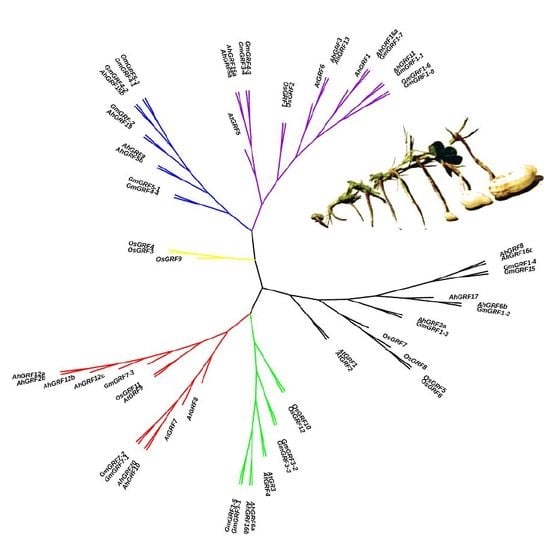
2.2. Genes Structure, Conserved Domains and Phylogenetic Analysis of AhGRF
2.3. Differential Expression Analysis of AhGRF Genes
2.4. Responses of AhGRFs to Exogenous GA3 Treatment
3. Discussion
4. Materials and Methods
4.1. Analysis of Peanut AhGRF Gene Family Members
4.2. AhGRF Gene Structure and Phylogenetic Analysis
4.3. Expression Profiling Based on RNA Sequencing (RNA-Seq) Data
4.4. Plant Materials and Hormone Treatments
4.5. Total RNA Extraction and cDNA Synthesis
4.6. Real-Time Quantitative PCR
4.7. Construction of AhGRF Transient Expression Vectors and Subcellular Localization Studies in Tobacco
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jeong, H.K.; Hirokazu, T. Regulation of plant growth and development by the growth-regulating factor and grf-interacting factor duo. J. Exp. Bot. 2015, 66, 6093–6107. [Google Scholar]
- Knaap, E.V.D.; Kim, H.; Kende, H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. J. Plant Physiol. 2000, 122, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Choi, D.; Kende, H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in arabidopsis. Plant J. 2003, 36, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Kim, J.H.; Kende, H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). J. Plant Cell Physiol. 2004, 45, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.F.; Li, B.; Jia, G.Q.; Zhang, T.F.; Dai, J.R.; Li, J.S.; Wang, S.C. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). J. Plant Sci. 2008, 175, 809–817. [Google Scholar] [CrossRef]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus, L.). J. Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ge, L.L.; Li, G.H.; Jiang, L.W.; Yang, Y.G. Characterization and expression analysis of growth regulating factor (GRF) family genes in cucumber. J. Arch. Biol. Sci. 2018, 70, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Roeber, B. Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. J. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Ertuğrul, F.L.Z.; İbrAhim, K.; Hüseyin, T. Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: In silico approaches. J. Turk. J. Biol. 2017, 38, 296–306. [Google Scholar]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. J. Plant Physiol. 2012, 159, 321–335. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Yang, H.L.; Zhan, G.M.; Li, R.J.; Deng, L.B.; Wang, X.F.; Liu, G.H.; Wang, H.Z. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 2012, 63, 3727–3740. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D.Y. Molecular Mechanism of microRNA396 Mediating Pistil Development in Arabidopsis. J. Plant Physiol. 2013, 164, 249–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vercruyssen, L.; Tognetti, V.B.; Gonzalez, N.; Van, D.J.; De, M.L.; Bielach, A.; De, R.R.; Van, B.F.; Inzé, D. Growth Regulating Factor 5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. J. Plant Physiol. 2015, 167, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. MiR396a-Mediated basic helix-loop-helix transcription factor bHLH74 Repression Acts as a Regulator for Root Growth in Arabidopsis Seedlings. J. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, B.H.; Jung, J.H.; Park, S.K.; Song, J.T.; Kim, J.H. Growth-regulating factor and grf-interacting factor specify meristematic cells of gynoecia and anthers. J. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, G.; Kim, G.T.; Tsukaya, H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005, 43, 68–78. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Fujikura, U.; Olas, J.J.; Xue, G.P.; Balazadeh, S.; Mueller-Roeber, B. Growth-Regulating Factor 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. J. PLoS Genet. 2018, 14, 1007484. [Google Scholar] [CrossRef]
- Nelissen, H.; Eechout, D.; Demuynck, K.; Persiau, G.; Walton, A.; van Bel, M.; Vervoort, M.; Candaele, J.; De Block, J.; De Jaeger, G. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 2015, 27, 1605–1619. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, D.; Xue, M.; Qian, J.; He, Y.; Wang, S. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 2014, 56, 1053–1063. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, B.H. Growth-regulating factor4ofarabidopsis thalianais required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 2006, 49, 463–468. [Google Scholar] [CrossRef]
- Daele, I.V.; Gonzalez, N.; Vercauteren, I.; Smet, L.D.; Inzé, D.; Roldan-Ruiz, I.; Vuylsteke, M. A comparative study of seed yield parameters in Arabidopsis thaliana mutants and transgenics. Plant Biotechnol. J. 2012, 10, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhang, W.; Wang, Y.; He, Q.; Shu, F.; Liu, H.; Wang, J.; Wang, J.; Yuan, L.; Deng, H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016, 58, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. J. Nat. Plants 2015, 203. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Wang, J.; Ju, Z.; Liu, Q.; Li, S.; Tian, H.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. J. Plant Sci. 2016, 247, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 214, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S.S.; Rake, A.V.; Miksche, J. Reassociation kinetics and cytophotometric characterization of peanut (Arachis hypogaea L.) DNA. J. Plant Physiol. 1980, 65, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Ozias-Akins, P. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Schmutz, J. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Yin, D.M.; Ji, C.M.; Ma, X.L.; Li, H.; Zhang, W.K.; Li, S.; Liu, F.Y.; Zhao, K.K.; Li, F.P.; Li, K.; et al. Genome of an allotetraploid wild peanut Arachis monticola: A de novo assembly. GigaScience 2018, 7. [Google Scholar] [CrossRef]
- Zhuang, W.J.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Varshney, R.K. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Yin, D.M.; Hu, X.F.; Zhang, X.G.; Sun, J.J.; Wang, Y.; Cui, D.Q. Temporal and spatial expression of oleate desaturase gene with different O/L values in peanut varieties. Chin. J. Oil Crop Sci. 2013, 35, 137–141. [Google Scholar]
- Wang, Y.; Zhang, X.G.; Zhao, M.Y.; Prakash, C.S.; He, G.; Yin, D. Insights into the novel members of the FAD2 gene family involved in high-oleate fluxes in peanut. J. Genome 2015, 58, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.E.; Mecchia, M.A.; Debernardi, J.M.; Schommer, C.; Weigel, D.; Palatnik, J.F. Control of cell proliferation in Arabidopsis thaliana by microrna mir396. J. Dev. 2010, 137, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of\r, GRF\r, transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Chu, C. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2015, 2, 15195. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in Rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.L.; Zhang, X.G.; Zhao, K.K.; Li, F.P.; Li, K.; Ning, L.L.; He, J.L.; Xin, Z.Y.; Yin, D.M. Small RNA and Degradome Deep Sequencing Reveals the Roles of microRNAs in Seed Expansion in Peanut (Arachis hypogaea L.). Front. Plant Sci. 2018, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Kuijt, S.J.H.; Greco, R.; Agalou, A.; Shao, J.; Hoen, C.C.; Overnäs, E.; Osnato, M.; Curiale, S.; Meynard, D.; Ouwerkerk, P.B. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX Families of Transcription Factors. Plant Physiol. 2014, 164, 1952–1966. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qiu, N.; Ding, Q.; Li, J.; Zhang, Y.; Li, H.; Cao, J. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2014, 15, 807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Z.; Jin, J.; Xie, X.; Zhang, H.; Chen, Q.; Luo, Z.; Yang, J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 2017, 639, 117–127. [Google Scholar] [CrossRef]
- Sparkes, I.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.J.; Li, J.B. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. J. BMC Genom. 2019, 20, 1471–2164. [Google Scholar]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Li, K.; Ning, L.; He, J.; Ma, X.; Li, Z.; Zhang, X.; Yin, D. Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 20, 4120. https://doi.org/10.3390/ijms20174120
Zhao K, Li K, Ning L, He J, Ma X, Li Z, Zhang X, Yin D. Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences. 2019; 20(17):4120. https://doi.org/10.3390/ijms20174120
Chicago/Turabian StyleZhao, Kunkun, Ke Li, Longlong Ning, Jialin He, Xingli Ma, Zhongfeng Li, Xingguo Zhang, and Dongmei Yin. 2019. "Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.)" International Journal of Molecular Sciences 20, no. 17: 4120. https://doi.org/10.3390/ijms20174120
APA StyleZhao, K., Li, K., Ning, L., He, J., Ma, X., Li, Z., Zhang, X., & Yin, D. (2019). Genome-Wide Analysis of the Growth-Regulating Factor Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences, 20(17), 4120. https://doi.org/10.3390/ijms20174120