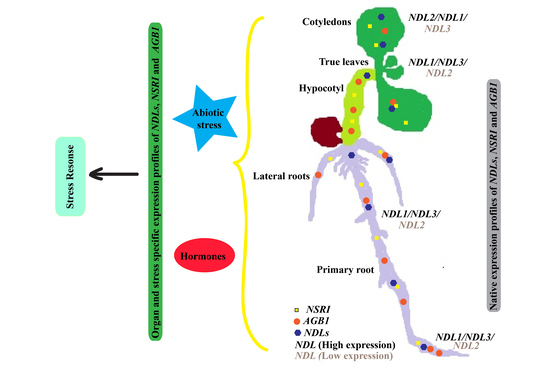
Arabidopsis NDL-AGB1 modules Play Role in Abiotic Stress and Hormonal Responses Along with Their Specific Functions
Abstract
:1. Introduction
- In silico comparative account of the regulatory elements and expression profiles of NDL1, NDL2, and NDL3 to ascertain their characteristics, similarities, and differences.
- In planta comparative analysis of the all three members of the NDL gene family during different stages of plant growth and development
- Comparative expression profile of NDL members in response to various abiotic stresses and hormonal treatments in presence and absence of AGB1.
2. Results
2.1. In Silico Analysis of the Upstream Regulatory Regions of Arabidopsis NDL1, NDL2 and NDL3
2.2. Comparative In Silico Expression Analysis of NDL Members
2.3. Comparative In Vivo Expression Analysis of NDLs during Early Stages of the Plant Growth
2.4. Comparative In Vivo Expression Analysis of NDLs in Absence of AGB1
2.5. In Silico Expression Analysis of NDLs under Various Stress and Hormonal Treatments
2.6. In Vivo Expression Analysis under Abiotic Stresses and Hormonal Treatments
2.7. In Vivo Expression Analysis of NDLs in Absence of AGB1 under Abiotic Stress and Hormonal Treatments
3. Discussion
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. Isolation and Cloning of NDL2 and NDL3 Promoters
4.3. In-Silico Analysis
4.4. GUS Staining Assay
4.5. Fluorometric GUS Assay
4.6. Hormone and Abiotic Stress Treatments
4.7. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Colaneri, A.C.; Tunc-Ozdemir, M.; Huang, J.P.; Jones, A.M. Growth attenuation under saline stress is mediated by the heterotrimeric G protein complex. BMC Plant Biol. 2014, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Ming, C.H.; Xu, D.B.; Fang, G.N.; Wang, E.H.; Gao, S.Q.; Xu, Z.S.; Li, L.C.; Zhang, X.H.; Miin, D.H. G-protein β subunit AGB1 positively regulates salt stress tolerance in Arabidopsis. J. Integr. Agric. 2015, 14, 314–325. [Google Scholar]
- Klopffleisch, K.; Phan, N.; Augustin, K.; Bayne, R.S.; Booker, K.S.; Botella, J.R.; Carpita, N.C.; Carr, T.; Chen, J.G.; Cooke, T.R.; et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 2011, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Kalaydjieva, L.; Gresham, D.; Gooding, R.; Heather, L.; Baas, F.; De Jonge, R.; Blechschmidt, K.; Angelicheva, D.; Chandler, D.; Worsley, P.; et al. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy–Lom. Am. J. Hum. Genet. 2000, 67, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.F.; Lu, J.; Zou, Y.S.; Soh-Won, J.; Cohen, D.M.; Buttrick, P.M.; Cooper, D.R.; Steinberg, S.F.; Mackman, N.; Pinsky, D.J.; et al. Hypoxia-associated induction of early growth response-1 gene expression. J. Biol. Chem. 1999, 274, 15030–15040. [Google Scholar] [CrossRef] [PubMed]
- Salnikow, K.; Su, W.; Blagosklonny, M.V.; Costa, M. Carcinogenic metals induce hypoxia-inducible factor-stimulated transcription by reactive oxygen species-independent mechanism. Cancer Res. 2000, 60, 3375–3378. [Google Scholar] [PubMed]
- Lachat, P.; Shaw, P.; Gebhard, S.; Van Belzen, N.; Chaubert, P.; Bosman, F.T. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem. Cell Biol. 2002, 118, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Salnikow, K.; Kluz, T.; Costa, M.; Piquemal, D.; Demidenko, Z.N.; Xie, K.; Blagosklonny, M.V. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol. Cell. Boil. 2002, 22, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhau, H.E.; Huang, W.C.; Iqbal, S.; Habib, F.K.; Sartor, O.; Cvitanovic, L.; Marshall, F.F.; Xu, Z.; Chung, L.W.K. cAMP-responsive element-binding protein regulates vascular endothelial growth factor expression: Implication in human prostate cancer bone metastasis. Oncogene 2007, 26, 5070–5077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Salnikow, K.; Costa, M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998, 58, 2182–2189. [Google Scholar] [PubMed]
- Krauter-Canham, R.; Bronner, R.; Evrard, J.L.; Hahne, G.; Friedt, W.; Steinmetz, A. A transmitting tissue-and pollen-expressed protein from sunflower with sequence similarity to the human RTP protein. Plant Sci. 1997, 129, 191–202. [Google Scholar] [CrossRef]
- Lazarescu, E.; Friedt, W.; Horn, R.; Steinmetz, A. Expression analysis of the sunflower SF21 gene family reveals multiple alternative and organ-specific splicing of transcripts. Gene 2006, 374, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Lazarescu, E.; Friedt, W.; Steinmetz, A. Organ-specific alternatively spliced transcript isoforms of the sunflower SF21C gene. Plant Cell Rep. 2010, 29, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, Y.; Uhrig, J.F.; Zhou, J.; Temple, B.; Jiang, K.; Jones, A.M. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein–mediated pathway. Plant Cell. 2009, 21, 3591–3609. [Google Scholar] [CrossRef] [PubMed]
- Rymaszewski, W.; Vile, D.; Bediee, A.; Dauzat, M.; Luchaire, N.; Kamrowska, D.; Granier, C.; Hennig, J. Stress-related gene expression reflects morphophysiological responses to water deficit. Plant Physiol. 2017, 174, 1913–1930. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, Y.; Ghawana, S.; Jones, A.M. N-MYC down-regulated-like proteins regulate meristem initiation by modulating auxin transport and MAX2 expression. PLoS ONE 2013, 8, e77863. [Google Scholar] [CrossRef] [PubMed]
- Luscher, B.; Eisenman, R.N. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990, 4, 2235–2241. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003, 33, 259–270. [Google Scholar] [CrossRef]
- Du, H.; Huang, F.; Wu, N.; Li, X.; Hu, H.; Xiong, L. Integrative regulation of drought escape through ABA-dependent and-independent pathways in rice. Mol. Plant. 2018, 4, 584–597. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Pan, Q.; Chen, S.; Feng, C.; Hai, J.; Li, H. Comparison of Trihelix transcription factors between wheat and Brachypodiumdistachyon at genome-wide. BMC Genom. 2019, 20, 142. [Google Scholar] [CrossRef]
- Rouached, H.; Secco, D.; Arpat, B.; Poirier, Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011, 11, 19. [Google Scholar] [CrossRef]
- Sobkowiak, L.; Bielewicz, D.; Małecka, E.; Jakobsen, I.; Albrechtsen, M.; Szweykowska-Kulinska, Z.; Pacak, A.M. The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front. Plant Sci. 2012, 3, 58. [Google Scholar] [CrossRef]
- Helin, K. Regulation of cell proliferation by the E2F transcription factors. Curr. Opin. Genet. Dev. 1998, 8, 28–35. [Google Scholar] [CrossRef]
- Lammens, T.; Li, J.; Leone, G.; De Veylder, L. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol. 2009, 19, 111–118. [Google Scholar] [CrossRef]
- De Veylder, L.; Beeckman, T.; Inze, D. The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 655. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Hattori, T.; Totsuka, M.; Hobo, T.; Kagaya, Y.; Yamamoto-Toyoda, A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002, 43, 136–140. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.N.; Xue, L.J.; Zou, M.J.; Liu, J.Y.; Chen, F.; Xue, H.W. Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 2011, 156, 1397–1409. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef]
- Skriver, K.; Olsen, F.L.; Rogers, J.C.; Mundy, J. Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 1991, 88, 7266–7270. [Google Scholar] [CrossRef]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Boter, M.; Ruiz-Rivero, O.; Abdeen, A.; Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004, 18, 1577–1591. [Google Scholar] [CrossRef]
- Kaplan, B.; Davydov, O.; Knight, H.; Galon, Y.; Knight, M.R.; Fluhr, R.; Fromm, H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis-elements in Arabidopsis. Plant Cell. 2006, 18, 2733–2748. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M.A. Role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032. [Google Scholar] [CrossRef]
- Yang, J.H.; Wang, H. Molecular mechanisms for vascular development and secondary cell wall formation. Front. Plant Sci. 2016, 7, 356. [Google Scholar] [CrossRef]
- Huang, T.; Harrar, Y.; Lin, C.; Reinhart, B.; Newell, N.R.; Talavera-Rauh, F.; Hokin, S.A.; Barton, M.K.; Kerstetter, R.A. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell. 2014, 26, 246–262. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Izhaki, A.; Bowman, J.L. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell. 2007, 19, 495–508. [Google Scholar] [CrossRef]
- Footitt, S.; Douterelo-Soler, I.; Clay, H.; Finch-Savage, W.E. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 20236–20241. [Google Scholar] [CrossRef]
- Xu, D.B.; Chen, M.; Ma, Y.N.; Xu, Z.S.; Li, L.C.; Chen, Y.F.; Ma, Y.Z.A. G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS ONE 2015, 10, e0116385. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- Chang, W.C.; Lee, T.Y.; Huang, H.D.; Huang, H.Y.; Pan, R.L. PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genom. 2008, 9, 561. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Bradford, M.M. ARapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ren, Y.; Zhao, J. Functional analysis of the rice metallothionein gene OsMT2b promoter in transgenic Arabidopsis plants and rice germinated embryos. Plant Sci. 2009, 176, 528–538. [Google Scholar] [CrossRef]










| S.NO. | TFs | Function |
|---|---|---|
| 1 | MYB1AT | Dehydration-responsive elements |
| 2 | ARR1AT | Cytokinin response regulators |
| 3 | GT1CONSENSUS | Salicylic acid-responsive elements, Light-responsive elements |
| 4 | MYCCONSENSUSAT | dehydration-responsive |
| 5 | SURECOREATSULTR11 | SURE contains auxin response factor (ARF) binding sequence |
| 6 | L1BOXATPDF1 | MYB binding motif |
| 7 | WBOXATNPR1 | Salicyclic acid-responsive elements |
| 8 | MYBCOREATCYCB1 | Cyclin B1-responsive elements |
| 9 | ABRELATERD1 | ABA-responsive elements |
| 10 | ANAERO1CONSENSUS | Anaerobic-responsive elements |
| 11 | WBBOXPCWRKY1 | Pathogenesis-related elements |
| 12 | LEAFYATAG | Target sequence of LEAFY |
| 13 | P1BS | Phosphate starvation-responsive elements |
| 14 | SORLIP1AT | Light-responsive elements |
| 15 | MYB1LEPR | Defence-related elements |
| S.NO. | TFs | Function |
|---|---|---|
| NDL1 | E2FAT | Cell cycle-responsive elements |
| RHERPATEXPA7 | Root hair specific-cis elements | |
| SBOXATRBCS | Sugar and ABA-responsive elements | |
| NDL2 | ABREATCONSENSUS | ABA-responsive elements |
| ABREATRD22 | ABA-responsive elements | |
| ABRERATCAL | Ca2+-responsive elements | |
| ACGTABREMOTIFA2OSEM | ABA-responsive elements | |
| GADOWNAT | GA-responsive elements | |
| MYBATRD22 | Dehydration-responsive elements | |
| SITEIIATCYTC | Responsible for oxidative phosphorylation | |
| T/GBOXATPIN2 | JA-responsive elements | |
| UP1ATMSD | Upregulation after main stem decapitation | |
| NDL3 | ABRERATCAL | Ca2+-responsive elements |
| AGL2ATCONSENSUS | AGAMOUS-LIKE 2 | |
| CGCGBOXAT | Ca2+/Calmodulin response elements | |
| GCCCORE | Pathogen-responsive elements |
| Col-0 | agb1-2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Stage | Organ | Tissue | AtNDL1 | AtNDL2 | AtNDL3 | AtNDL1 | AtNDL2 | AtNDL3 |
| 8-day old seedlings | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | ++ | − | − | ||
| MZ | ++ | ++ | − | − | ++ | − | ||
| LR | ++ | − | ++ | ++ | − | ++ | ||
| Hypocotyl | − | ++ | − | − | ++ | − | ||
| Cotyledons | ++ | ++ | − | − | ++ | − | ||
| Leaves | ++ | ++ | ++ | ++ | ++ | ++ | ||
| 6-day old seedlings (Cold) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | ++ | − | − | ||
| MZ | + | + | − | + | ++ | − | ||
| Hypocotyl | ++ | ++ | ++ | − | ++ | − | ||
| Cotyledons | +++ | +++ | ++ | − | ++ | − | ||
| 6-day old seedlings (Heat) | PR | RT | − | − | ++ | − | − | ++ |
| CDZ | − | − | ++ | − | − | ++ | ||
| EZ | − | − | − | − | − | − | ||
| MZ | − | ++ | − | − | ++ | − | ||
| Hypocotyl | + | ++ | − | − | ++ | − | ||
| Cotyledons | − | ++ | + | − | ++ | − | ||
| 6-day old seedlings (Mannitol) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | + | − | + | ++ | − | ++ | ||
| EZ | + | − | − | − | − | − | ||
| MZ | − | ++ | − | − | ++ | − | ||
| Hypocotyl | ++ | ++ | − | − | ++ | − | ||
| Cotyledons | ++ | ++ | − | + | ++ | + | ||
| 6-day old seedlings (PEG) | PR | RT | +++ | − | − | − | − | +++ |
| CDZ | +++ | − | − | +++ | − | ++ | ||
| EZ | ++ | − | − | + | − | − | ||
| MZ | − | ++ | − | − | ++ | − | ||
| Hypocotyl | − | ++ | + | − | ++ | − | ||
| Cotyledons | ++ | ++ | + | − | ++ | − | ||
| 6-day old seedlings (Salt) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | + | − | − | ++ | − | − | ||
| MZ | + | ++ | − | − | ++ | − | ||
| Hypocotyl | ++ | ++ | + | − | ++ | + | ||
| Cotyledons | ++ | ++ | + | − | ++ | − | ||
| 6-day old seedlings (ABA) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | ++ | − | − | ||
| MZ | − | ++ | − | ++ | ++ | − | ||
| Hypocotyl | − | ++ | − | − | ++ | − | ||
| Cotyledons | ++ | +++ | ++ | − | ++ | − | ||
| 6-day old seedlings (GA) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | +++ | − | − | ||
| MZ | + | + | − | ++ | ++ | − | ||
| Hypocotyl | + | ++ | + | − | ++ | − | ||
| Cotyledons | ++ | ++ | + | − | ++ | − | ||
| 6-day old seedlings (IAA) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | +++ | − | ++ | ++ | − | ++ | ||
| EZ | +++ | − | − | ++ | − | − | ||
| MZ | ++ | + | − | + | ++ | − | ||
| Hypocotyl | ++ | ++ | + | − | ++ | − | ||
| Cotyledons | ++ | ++ | ++ | − | ++ | − | ||
| 6-day old seedlings (JA) | PR | RT | ++ | − | ++ | +++ | − | ++ |
| CDZ | ++ | − | ++ | +++ | − | ++ | ||
| EZ | ++ | − | − | +++ | − | − | ||
| MZ | ++ | + | − | ++ | ++ | − | ||
| Hypocotyl | ++ | ++ | − | − | ++ | − | ||
| Cotyledons | ++ | +++ | − | − | ++ | − | ||
| 6-day old seedlings (SA) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | + | − | ++ | ||
| EZ | + | − | − | + | − | − | ||
| MZ | − | ++ | − | − | ++ | − | ||
| Hypocotyl | + | ++ | + | − | ++ | − | ||
| Cotyledons | ++ | ++ | + | − | ++ | + | ||
| 6-day old seedlings (BAP) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | ++ | − | − | ||
| MZ | + | ++ | − | ++ | ++ | − | ||
| Hypocotyl | − | ++ | − | − | ++ | − | ||
| Cotyledons | +++ | ++ | + | − | ++ | + | ||
| 6-day old seedlings (Control) | PR | RT | ++ | − | ++ | − | − | ++ |
| CDZ | ++ | − | ++ | ++ | − | ++ | ||
| EZ | ++ | − | − | ++ | − | − | ||
| MZ | − | ++ | − | ++ | ++ | − | ||
| Hypocotyl | − | ++ | ++ | − | ++ | − | ||
| Cotyledons | ++ | ++ | ++ | − | ++ | − | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katiyar, A.; Mudgil, Y. Arabidopsis NDL-AGB1 modules Play Role in Abiotic Stress and Hormonal Responses Along with Their Specific Functions. Int. J. Mol. Sci. 2019, 20, 4736. https://doi.org/10.3390/ijms20194736
Katiyar A, Mudgil Y. Arabidopsis NDL-AGB1 modules Play Role in Abiotic Stress and Hormonal Responses Along with Their Specific Functions. International Journal of Molecular Sciences. 2019; 20(19):4736. https://doi.org/10.3390/ijms20194736
Chicago/Turabian StyleKatiyar, Arpana, and Yashwanti Mudgil. 2019. "Arabidopsis NDL-AGB1 modules Play Role in Abiotic Stress and Hormonal Responses Along with Their Specific Functions" International Journal of Molecular Sciences 20, no. 19: 4736. https://doi.org/10.3390/ijms20194736
APA StyleKatiyar, A., & Mudgil, Y. (2019). Arabidopsis NDL-AGB1 modules Play Role in Abiotic Stress and Hormonal Responses Along with Their Specific Functions. International Journal of Molecular Sciences, 20(19), 4736. https://doi.org/10.3390/ijms20194736