Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7
Abstract
:1. Introduction
2. Results
2.1. Purification and Folding of lncTCF7
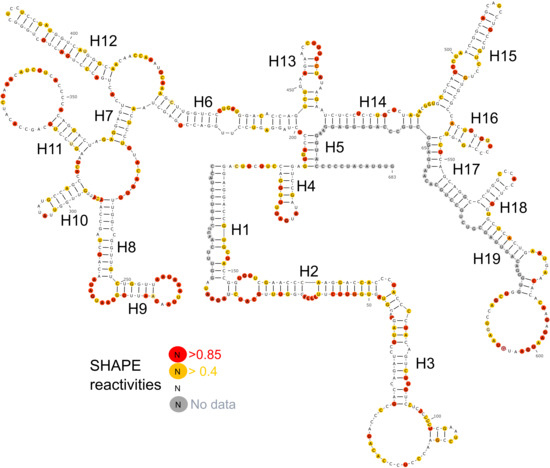
2.2. Determining the Secondary Structure of lncTCF7
2.2.1. Shotgun Secondary Structure Analysis
2.2.2. DMS Probing
2.2.3. Confidence Estimation
2.3. Identifying the Well-Defined Structures in lncTCF7
2.4. Identifying the Conserved Regions in lncTCF7
3. Discussion
4. Materials and Methods
4.1. Plasmids and DNA Templates
4.2. RNA Synthesis and Purification
4.3. Chemical Probing
4.3.1. SHAPE-MaP
4.3.2. DMS-MaP
4.3.3. 3S Shotgun Secondary Structure Analysis
4.4. Structure Determination and Confidence Estimation
4.5. Shannon Entropy Calculation
4.6. Sequence and Structure Conservation Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| lncRNA | Long non-coding RNA |
| SHAPE | Selective 2′-Hydroxyl Acylation analyzed by Primer Extension |
| DMS | Dimethyl Sulfate |
| SEC | Size Exclusion Chromatography |
| WSPAR | WNT Signaling Pathway Activating Non-Coding RNA |
| SRA | Steroid Receptor RNA Activator |
| HOTAIR | Hox Transcript Antisense Interfering RNA |
| TCF7 | Transcription Factor 7 |
| SWI/SNF | Switch/Sucrose Non-Fermentable |
| 3S | Shotgun Secondary Structure |
| MaP | Mutational Profiling |
| R-scape | RNA Structural Conservation Above Phylogenetic Expectation |
| CNBP | Cellular Nucleic Acid-Binding Protein |
| 1M7 | 1-methyl-7-nitroisatoic anhydride |
| DMSO | Dimethyl Sulfoxide |
References
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.R.; Feng, F.Y.; Chinnaiyan, A.M. The bright side of dark matter: lncRNAs in cancer. J. Clin. Investig. 2016, 126, 2775–2782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb. Perspect. Med. 2017, 7, a026492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2013, 15, 7. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012, 41, D983–D986. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, P.; Wang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Li, X.; Fang, Y.; Shen, W.; Xu, Y.; et al. Lnc2Cancer v2.0: Updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2018, 47, D1028–D1033. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2018, 47, D135–D139. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Lodish, H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 2017, 130, 1965. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Sizing up long non-coding RNAs: Do lncRNAs have secondary and tertiary structure? BioArchitecture 2012, 2, 189–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Martens, L.; Rühle, F.; Stoll, M. LncRNA secondary structure in the cardiovascular system. Non-Coding RNA Res. 2017, 2, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Hennelly, S.; Doyle, B.; Gulati, A.A.; Novikova, I.V.; Sanbonmatsu, K.Y.; Boyer, L.A. A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage. Mol. Cell 2016, 64, 37–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Mao, Y.S.; Diermeier, S.D.; Novikova, I.V.; Nawrocki, E.P.; Jones, T.A.; Lazar, Z.; Tung, C.-S.; Luo, W.; Eddy, S.R.; et al. Identification and Characterization of a Class of MALAT1-like Genomic Loci. Cell Rep. 2017, 19, 1723–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012, 40, 5034–5051. [Google Scholar] [CrossRef] [PubMed]
- Somarowthu, S.; Legiewicz, M.; Chillon, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef]
- Liu, F.; Somarowthu, S.; Pyle, A.M. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017, 13, 282–289. [Google Scholar] [CrossRef]
- Sherpa, C.; Rausch, J.W.; Le Grice, S.F. Structural characterization of maternally expressed gene 3 RNA reveals conserved motifs and potential sites of interaction with polycomb repressive complex 2. Nucleic Acids Res. 2018, 46, 10432–10447. [Google Scholar] [CrossRef] [Green Version]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.-M.; Pellequer, J.-L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chen, M.; Gao, M.; Cong, Y.; Jiang, L.; Wei, J.; Huang, J. Down-regulation of lncTCF7 inhibits cell migration and invasion in colorectal cancer via inhibiting TCF7 expression. Hum. Cell 2019, 32, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Liang, X.L.; Wu, Y.F.; Pang, Y.H.; Zhao, X.J.; Lu, Y.X. ILK promotes survival and self-renewal of hypoxic MSCs via the activation of lncTCF7-Wnt pathway induced by IL-6/STAT3 signaling. Gene Ther. 2019, 26, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, H.L. LncRNA TCF7 triggered endoplasmic reticulum stress through a sponge action with miR-200c in patients with diabetic nephropathy. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5912–5922. [Google Scholar] [PubMed]
- Li, T.; Zhu, J.; Zuo, S.; Chen, S.; Ma, J.; Ma, Y.; Guo, S.; Wang, P.; Liu, Y. 1,25(OH)2D3 Attenuates IL-1beta-Induced Epithelial-to-Mesenchymal Transition Through Inhibiting the Expression of lncTCF7. Oncol. Res. 2019, 27, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Xu, J.; Xiao, Q.; Chen, F.; Han, X. Long non-coding RNA TCF7 contributes to the growth and migration of airway smooth muscle cells in asthma through targeting TIMMDC1/Akt axis. Biochem. Biophys. Res. Commun. 2019, 508, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, L.; Zheng, L.; Hong, Y.; Zhao, L. LncRNATCF7 promotes the growth and self-renewal of glioma cells via suppressing the miR-200c-EpCAM axis. Biomed. Pharmacother. 2018, 97, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.S.; Wang, H.M.; Song, X.Y. Long non-coding RNA TCF7 predicts the progression and facilitates the growth and metastasis of colorectal cancer. Mol. Med. Rep. 2018, 17, 6902–6908. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wang, D. Long noncoding RNA TCF7 promotes invasiveness and self-renewal of human non-small cell lung cancer cells. Hum. Cell 2017, 30, 23–29. [Google Scholar] [CrossRef]
- Li, T.; Zhu, J.; Wang, X.; Chen, G.; Sun, L.; Zuo, S.; Zhang, J.; Chen, S.; Ma, J.; Yao, Z.; et al. Long non-coding RNA lncTCF7 activates the Wnt/beta-catenin pathway to promote metastasis and invasion in colorectal cancer. Oncol. Lett. 2017, 14, 7384–7390. [Google Scholar]
- Gao, X.; Guo, X.; Xue, H.; Qiu, W.; Guo, X.; Zhang, J.; Qian, M.; Li, T.; Liu, Q.; Shen, J.; et al. lncTCF7 is a negative prognostic factor, and knockdown of lncTCF7 inhibits migration, proliferation and tumorigenicity in glioma. Sci. Rep. 2017, 7, 17456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Zhang, J.; Shen, B.; Yin, K.; Xu, J.; Gao, W.; Zhang, L. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. 2015, 34, 116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; He, L.; Du, Y.; Zhu, P.; Huang, G.; Luo, J.; Yan, X.; Ye, B.; Li, C.; Xia, P.; et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015, 16, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Chillon, I.; Marcia, M.; Legiewicz, M.; Liu, F.; Somarowthu, S.; Pyle, A.M. Native Purification and Analysis of Long RNAs. Methods Enzymol. 2015, 558, 3–37. [Google Scholar] [PubMed] [Green Version]
- Pyle, A. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002, 7, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Draper, D.E. On the role of magnesium ions in RNA stability. Biopolymers 1998, 48, 113–135. [Google Scholar] [CrossRef]
- Woodson, S.A. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005, 9, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Low, J.T.; Weeks, K.M. SHAPE-directed RNA secondary structure prediction. Methods 2010, 52, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Smola, M.J.; Rice, G.M.; Busan, S.; Siegfried, N.A.; Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015, 10, 1643–1669. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Tackling structures of long noncoding RNAs. Int. J. Mol. Sci. 2013, 14, 23672–23684. [Google Scholar] [CrossRef]
- Novikova, I.V.; Dharap, A.; Hennelly, S.P.; Sanbonmatsu, K.Y. 3S: Shotgun secondary structure determination of long non-coding RNAs. Methods 2013, 63, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Tijerina, P.; Mohr, S.; Russell, R. DMS footprinting of structured RNAs and RNA—Protein complexes. Nat. Protoc. 2007, 2, 2608. [Google Scholar] [CrossRef] [PubMed]
- Zubradt, M.; Gupta, P.; Persad, S.; Lambowitz, A.M.; Weissman, J.S.; Rouskin, S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 2016, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Ding, F.; Weeks, K.M.; Dokholyan, N.V. Statistical Analysis of SHAPE-Directed RNA Secondary Structure Modeling. Biochemistry 2013, 52, 596–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathews, D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA 2004, 10, 1178–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dethoff, E.A.; Weeks, K.M. Effects of Refolding on Large-Scale RNA Structure. Biochemistry 2019, 58, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 3692–3697. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Rivas, E.; Clements, J.; Eddy, S.R. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods 2016, 14, 45. [Google Scholar] [CrossRef]
- Harmanci, A.O.; Sharma, G.; Mathews, D.H. TurboFold: Iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinform. 2011, 12, 108. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300. [Google Scholar] [CrossRef] [PubMed]
- Portoso, M.; Ragazzini, R.; Brenčič, Ž.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.C.A.; Pyle, A.M.; Somarowthu, S. Phylogenetic Analysis with Improved Parameters Reveals Conservation in lncRNA Structures. J. Mol. Biol. 2019, 431, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Busan, S.; Weeks, K.M. Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 2018, 24, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Karabiber, F.; McGinnis, J.L.; Favorov, O.V.; Weeks, K.M. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013, 19, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Giardine, B.; Riemer, C.; Hardison, R.C.; Burhans, R.; Elnitski, L.; Shah, P.; Zhang, Y.; Blankenberg, D.; Albert, I.; Taylor, J.; et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005, 15, 1451–1455. [Google Scholar] [CrossRef] [Green Version]
- Blanchette, M.; Kent, W.J.; Riemer, C.; Elnitski, L.; Smit, A.F.A.; Roskin, K.M.; Baertsch, R.; Rosenbloom, K.; Clawson, H.; Green, E.D.; et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004, 14, 708–715. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Owens, M.C.; Clark, S.C.; Yankey, A.; Somarowthu, S. Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7. Int. J. Mol. Sci. 2019, 20, 4770. https://doi.org/10.3390/ijms20194770
Owens MC, Clark SC, Yankey A, Somarowthu S. Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7. International Journal of Molecular Sciences. 2019; 20(19):4770. https://doi.org/10.3390/ijms20194770
Chicago/Turabian StyleOwens, Michael C., Sean C. Clark, Allison Yankey, and Srinivas Somarowthu. 2019. "Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7" International Journal of Molecular Sciences 20, no. 19: 4770. https://doi.org/10.3390/ijms20194770
APA StyleOwens, M. C., Clark, S. C., Yankey, A., & Somarowthu, S. (2019). Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7. International Journal of Molecular Sciences, 20(19), 4770. https://doi.org/10.3390/ijms20194770