Expression Profile of Cell Cycle-Related Genes in Human Fibroblasts Exposed Simultaneously to Radiation and Simulated Microgravity
Abstract
:1. Introduction
2. Results
2.1. Gene Expression Profile Changes after Radiation and/or Simulated μG Treatment
2.2. Radiation Exposure Led to a Marked Change in the Cell Cycle-Related Gene Expression Profile
2.3. Changes in Cell Cycle-Related Genes Expression Profile in Cells under Simulated μG and Radiation
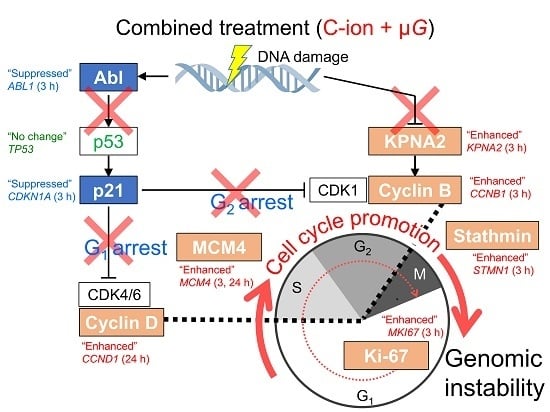
3. Discussion
3.1. Gene Expression Profile Changes with Simulated μG
3.2. X-ray and C-Ion Irradiation Induced Changes in Gene Expression
3.3. Synergistic Effect of Radiation and Simulated μG on Changes in Expression Profile of Cell Cycle-Related Genes
4. Materials and Methods
4.1. Cell Culture
4.2. Synchronized Irradiation Systems Under Simulated μG or 1G
4.3. Experimental Design
4.4. RNA Extraction
4.5. RNA Sequencing
4.6. Pathway Analysis
4.7. Heatmap Representation for Visualization of Changing Gene Expression Level
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | Three-dimensional |
| ABL1 | ABL proto-oncogene 1, non-receptor tyrosine kinase (c-Abl) |
| ACTA1 | Actin alpha 1, skeletal muscle |
| AEC | Accelerator Engineering Co. |
| AES | Advanced Engineering Services Co. Ltd. |
| ATM | ATM serine/threonine kinase |
| CCNB1 | Cyclin B1 |
| CCND1 | Cyclin D1 |
| CDC20 | Cell division cycle 20 |
| CDK1 | Cyclin-dependent kinase 1 (= cell division cycle protein 2, cdc2) |
| CDK4/6 | Cyclin-dependent kinase 4/6 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (= p21) |
| C-ion | Carbon ion |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| DCC | Disposable sealed irradiation cell culture chamber |
| EDGE | Empirical Analysis of DGE |
| GHMC | Gunma University Heavy Ion Medical Center |
| GIAR | Gunma University Initiative for Advanced Research |
| ISS | International Space Station |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KPNA2 | Karyopherin subunit alpha 2 |
| LET | Linear energy transfer |
| MCM4 | Minichromosome maintenance complex component 4 |
| MDM2 | MDM2 proto-oncogene |
| MeV/n | Megaelectronvolt per nucleon |
| μG | Microgravity |
| MKI67 | Marker of proliferation Ki-67 |
| NIAID | National Institute of Allergy and Infectious Diseases |
| NIH | National Institutes of Health |
| p53 | p53 tumor suppressor protein |
| RO | Rotation for simulated μG |
| ST | Standing 1G |
| STMN1 | Stathmin 1 |
| TFDP1 | Transcription factor Dp-1 |
References
- Kahn, J.; Liverman, C.T.; McCoy, M.A. Committee on Ethics Principles and Guidelines for Health Standards for Long Duration and Exploration Spaceflights. In Health Standards for Long Duration and Exploration Spaceflight: Ethics Principles, Responsibilities, and Decision Framework; National Academies Press: Washington, DC, USA, 2014; ISBN 978-0-309-29657-1. [Google Scholar]
- Cucinotta, F.A. A new approach to reduce uncertainties in space radiation cancer risk predictions. PLoS ONE 2015, 10, e0120717. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A. Space radiation risks for astronauts on multiple International Space Station missions. PLoS ONE 2014, 9, e96099. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Manuel, F.K.; Jones, J.; Iszard, G.; Murrey, J.; Djojonegro, B.; Wear, M. Space radiation and cataracts in astronauts. Radiat. Res. 2001, 156, 460–466. [Google Scholar] [CrossRef]
- Jones, J.A.; McCarten, M.; Manuel, K.; Djojonegoro, B.; Murray, J.; Feiversen, A.; Wear, M. Cataract formation mechanisms and risk in aviation and space crews. Aviat. Space Environ. Med. 2007, 78, A56–A66. [Google Scholar] [PubMed]
- Cucinotta, F.A.; Alp, M.; Sulzman, F.M.; Wang, M. Space radiation risks to the central nervous system. Life Sci. Space Res. 2014, 2, 54–69. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Gonzalo, R.; Baatout, S.; Moreels, M. Impact of particle irradiation on the immune system: From the clinic to Mars. Front. Immunol. 2017, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Delp, M.D.; Charvat, J.M.; Limoli, C.L.; Globus, R.K.; Ghosh, P. Apollo lunar astronauts show higher cardiovascular disease mortality: Possible deep space radiation effects on the vascular endothelium. Sci. Rep. 2016, 6, 29901. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Hamada, N.; Little, M.P. No evidence for an increase in circulatory disease mortality in astronauts following space radiation exposures. Life Sci. Space Res. 2016, 10, 53–56. [Google Scholar] [CrossRef]
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef]
- Bender, M.A.; Gooch, P.C.; Kondo, S. The Gemini-XI S-4 spaceflight-radiation interaction experiment: The human blood experiment. Radiat. Res. 1968, 34, 228–238. [Google Scholar] [CrossRef]
- Horneck, G.; Rettberg, P.; Kozubek, S.; Baumstark-Khan, C.; Rink, H.; Schäfer, M.; Schmitz, C. The influence of microgravity on repair of radiation-induced DNA damage in bacteria and human fibroblasts. Radiat. Res. 1997, 147, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Pross, H.D.; Kiefer, J. Repair of cellular radiation damage in space under microgravity conditions. Radiat. Environ. Biophys. 1999, 38, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Ohnishi, K.; Takahashi, S.; Masukawa, M.; Sekikawa, K.; Amano, T.; Nakano, T.; Nagaoka, S.; Ohnishi, T. The effects of microgravity on induced mutation in Escherichia coli and Saccharomyces cerevisiae. Adv. Space Res. 2001, 28, 555–561. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohnishi, K.; Takahashi, S.; Masukawa, M.; Sekikawa, K.; Amano, T.; Nakano, T.; Nagaoka, S.; Ohnishi, T. The effects of microgravity on ligase activity in the repair of DNA double-strand breaks. Int. J. Radiat. Biol. 2000, 76, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Bücker, H.; Horneck, G.; Reitz, G. Embryogenesis and organogenesis of Carausius morosus under spaceflight conditions. Naturwissenschaften 1986, 73, 433–434. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xu, D.; Zhao, L.; Zhang, M.; Sun, Y. Effects of microgravity on DNA damage response in Caenorhabditis elegans during Shenzhou-8 spaceflight. Int. J. Radiat. Biol. 2015, 91, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Ikenaga, M.; Yoshikawa, I.; Kojo, M.; Ayaki, T.; Ryo, H.; Ishizaki, K.; Kato, T.; Yamamoto, H.; Hara, R. Mutations induced in Drosophila during space flight. Biol. Sci. Space 1997, 11, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kikuchi, M.; Nagaoka, S.; Watanabe, H. Recovery of Deinococcus radiodurans from radiation damage was enhanced under microgravity. Biol. Sci. Space 1996, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Yatagai, F.; Ishioka, N. Are biological effects of space radiation really altered under the microgravity environment? Life Sci. Space Res. 2014, 3, 76–89. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Wong, M.; Lu, T.; Zhang, Y.; Wu, H. Interplay of space radiation and microgravity in DNA damage and DNA damage response. NPJ Microgravity 2017, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Yatagai, F.; Honma, M.; Dohmae, N.; Ishioka, N. Biological effects of space environmental factors: A possible interaction between space radiation and microgravity. Life Sci. Space Res. 2019, 20, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Indo, H.P.; Tomiyoshi, T.; Suenaga, S.; Tomita, K.; Suzuki, H.; Masuda, D.; Terada, M.; Ishioka, N.; Gusev, O.; Cornette, R.; et al. MnSOD downregulation induced by extremely low 0.1 mGy single and fractionated X-rays and microgravity treatment in human neuroblastoma cell line, NB-1. J. Clin. Biochem. Nutr. 2015, 57, 98–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Sun, Q.; Xu, W.; Li, F.; Li, H.; Lu, J.; Wu, L.; Wu, Y.; Liu, M.; Bian, P. Modulation of modeled microgravity on radiation-induced bystander effects in Arabidopsis thaliana. Mutat. Res. 2015, 773, 27–36. [Google Scholar] [CrossRef]
- Beck, M.; Moreels, M.; Quintens, R.; Abou-El-Ardat, K.; El-Saghire, H.; Tabury, K.; Michaux, A.; Janssen, A.; Neefs, M.; van Oostveldt, P.; et al. Chronic exposure to simulated space conditions predominantly affects cytoskeleton remodeling and oxidative stress response in mouse fetal fibroblasts. Int. J. Mol. Med. 2014, 34, 606–615. [Google Scholar] [CrossRef] [Green Version]
- Pani, G.; Verslegers, M.; Quintens, R.; Samari, N.; de Saint-Georges, L.; van Oostveldt, P.; Baatout, S.; Benotmane, M.A. Combined exposure to simulated microgravity and acute or chronic radiation reduces neuronal network integrity and survival. PLoS ONE 2016, 11, e0155260. [Google Scholar] [CrossRef]
- Herranz, R.; Anken, R.; Boonstra, J.; Braun, M.; Christianen, P.C.; de Geest, M.; Hauslage, J.; Hilbig, R.; Hill, R.J.; Lebert, M.; et al. Ground-based facilities for simulation of microgravity: Organism-specific recommendations for their use, and recommended terminology. Astrobiology 2013, 13, 1–17. [Google Scholar] [CrossRef]
- Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T.; Takahashi, A. A new system for three-dimensional clinostat synchronized X-irradiation with high-speed shutter for space radiation research. Biol. Sci. Space 2016, 30, 8–16. [Google Scholar] [CrossRef]
- Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T.; Takahashi, A. Development and performance evaluation of a three-dimensional clinostat synchronized heavy-ion irradiation system. Life Sci. Space Res. 2017, 12, 51–60. [Google Scholar] [CrossRef]
- Takahashi, A.; Ikeda, H.; Souda, H.; Puspitasari, A.; Held, K.D.; Hidema, J.; Nikawa, T.; Yoshida, Y.; Kanai, T. Performance evaluation of new devices to synchronize heavy-ion or X-ray irradiation and simulated microgravity for space radiation research~Improvement of a control stage. In Proceedings of the 31st International Symposium on Space Technology and Science, Tokyo, Japan, 6 June 2017. [Google Scholar]
- Hada, M.; Ikeda, H.; Rhone, J.R.; Beitman, A.J.; Plante, I.; Souda, H.; Yoshida, Y.; Held, K.D.; Fujiwara, K.; Saganti, P.B.; et al. Increased chromosome aberrations in cells exposed simultaneously to simulated microgravity and radiation. Int. J. Mol. Sci. 2019, 20, 43. [Google Scholar] [CrossRef]
- Hartwell, L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell 1992, 71, 543–546. [Google Scholar] [CrossRef]
- Cho, R.J.; Huang, M.; Campbell, M.J.; Dong, H.; Steinmetz, L.; Sapinoso, L.; Hampton, G.; Elledge, S.J.; Davis, R.W.; Lockhart, D.J. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001, 27, 48–54. [Google Scholar] [CrossRef]
- Cohen, B.A.; Mitra, R.D.; Hughes, J.D.; Church, G.M. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000, 26, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, C.; Hassler, D.M.; Cucinotta, F.A.; Ehresmann, B.; Wimmer-Schweingruber, R.F.; Brinza, D.E.; Kang, S.; Weigle, G.; Böttcher, S.; Böhm, E.; et al. Measurements of energetic particle radiation in transit to Mars on the Mars science laboratory. Science 2013, 340, 1080–1084. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Formisano, L.; Lu, Y.; Servetto, A.; Hanker, A.B.; Jansen, V.M.; Bauer, J.A.; Sudhan, D.R.; Guerrero-Zotano, A.L.; Croessmann, S.; Guo, Y.; et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 2018, 10, 1373. [Google Scholar] [CrossRef] [PubMed]
- Levav-Cohen, Y.; Goldberg, Z.; Zuckerman, V.; Grossman, T.; Haupt, S.; Haupt, Y. C-Abl as a modulator of p53. Biochem. Biophys. Res. Commun. 2005, 331, 737–749. [Google Scholar] [CrossRef]
- Charrier-Savournin, F.B.; Château, M.T.; Gire, V.; Sedivy, J.; Piette, J.; Dulic, V. p21-Mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol. Biol. Cell. 2004, 15, 3965–3976. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Elledge, S.J.; Keyomarsi, K.; Dynlacht, B.; Tsai, L.H.; Zhang, P.; Dobrowolski, S.; Bai, C.; Connell-Crowley, L.; Swindell, E. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell. 1995, 6, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.L.; Wang, G.W.; Yang, G.Q.; Yang, H.; Zhuang, L. Karyopherin subunit-α 2 expression accelerates cell cycle progression by upregulating CCNB2 and CDK1 in hepatocellular carcinoma. Oncol. Lett. 2018, 15, 2815–2820. [Google Scholar] [CrossRef]
- Ishimi, Y.; Komamura-Kohno, Y.; Kwon, H.J.; Yamada, K.; Nakanishi, M. Identification of MCM4 as a target of the DNA replication block checkpoint system. J. Biol. Chem. 2003, 278, 24644–24650. [Google Scholar] [CrossRef]
- Giono, L.E.; Resnick-Silverman, L.; Carvajal, L.A.; St Clair, S.; Manfredi, J.J. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase. Oncogene 2017, 36, 6762–6773. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Bizhanova, A.; Matheson, T.D.; Yu, J.; Zhu, L.J.; Kaufman, P.D. Ki-67 contributes to normal cell cycle progression and inactive x heterochromatin in p21 checkpoint-proficient human cells. Mol. Cell. Biol. 2017, 37, e00569-16. [Google Scholar] [CrossRef]
- Rubin, C.I.; Atweh, G.F. The role of stathmin in the regulation of the cell cycle. J. Cell. Biochem. 2004, 93, 242–250. [Google Scholar] [CrossRef]
- Hauslage, J.; Cevik, V.; Hemmersbach, R. Pyrocystis noctiluca represents an excellent bioassay for shear forces induced in ground-based microgravity simulators (clinostat and random positioning machine). npj Microgravity 2017, 3, 12. [Google Scholar] [CrossRef]
- Todd, P. Physical effects at the cellular level under altered gravity conditions. Adv. Space Res. 1992, 12, 43–49. [Google Scholar] [CrossRef]
- Deckbar, D.; Jeggo, P.A.; Löbrich, M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Jeggo, P.A.; Löbrich, M. Contribution of DNA repair and cell cycle checkpoint arrest to the maintenance of genomic stability. DNA Repair 2006, 5, 1192–1198. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Ohnishi, K.; Tsuji, K.; Matsumoto, H.; Aoki, H.; Wang, X.; Tamamoto, T.; Yukawa, O.; Furusawa, Y.; Ejima, Y.; et al. WAF1 accumulation by carbon-ion beam and alpha-particle irradiation in human glioblastoma cultured cells. Int. J. Radiat. Biol. 2000, 76, 335–341. [Google Scholar] [CrossRef]
- Yasuda, H.; Komiyama, T.; Fujitaka, K. Probability of hippocampus cell hits by high-LET space radiation in a low-Earth-orbit mission (STS-91). Phys. Med. 2001, 17, S166–S169. [Google Scholar]
- Wang, J.; Zhang, J.; Bai, S.; Wang, G.; Mu, L.; Sun, B.; Wang, D.; Kong, Q.; Liu, Y.; Yao, X.; et al. Simulated microgravity promotes cellular senescence via oxidant stress in rat PC12 cells. Neurochem. Int. 2009, 55, 710–716. [Google Scholar] [CrossRef]
- Coinu, R.; Chiaviello, A.; Galleri, G.; Franconi, F.; Crescenzi, E.; Palumbo, G. Exposure to modeled microgravity induces metabolic idleness in malignant human MCF-7 and normal murine VSMC cells. FEBS Lett. 2006, 580, 2465–2470. [Google Scholar] [CrossRef] [Green Version]
- Cotrupi, S.; Ranzani, D.; Maier, J.A. Impact of modeled microgravity on microvascular endothelial cells. Biochim. Biophys. Acta. 2005, 1746, 163–168. [Google Scholar] [CrossRef] [Green Version]
- Arase, Y.; Nomura, J.; Sugaya, S.; Sugita, K.; Kita, K.; Suzuki, N. Effects of 3-D clino-rotation on gene expression in human fibroblast cells. Cell Biol. Int. 2002, 26, 225–233. [Google Scholar] [CrossRef]
- Yuge, L.; Kajiume, T.; Tahara, H.; Kawahara, Y.; Umeda, C.; Yoshimoto, R.; Wu, S.L.; Yamaoka, K.; Asashima, M.; Kataoka, K.; et al. Microgravity potentiates stem cell proliferation while sustaining the capability of differentiation. Stem Cells Dev. 2006, 15, 921–929. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Pan, S.; Zhang, L.; Zhang, W.; Yi, H.; Niu, Y. Three-dimensional simulated microgravity culture improves the proliferation and odontogenic differentiation of dental pulp stem cell in PLGA scaffolds implanted in mice. Mol. Med. Rep. 2017, 15, 873–878. [Google Scholar] [CrossRef]
- Lei, X.H.; Ning, L.N.; Cao, Y.J.; Liu, S.; Zhang, S.B.; Qiu, Z.F.; Hu, H.M.; Zhang, H.S.; Liu, S.; Duan, E.K. NASA-approved rotary bioreactor enhances proliferation of human epidermal stem cells and supports formation of 3D epidermis-like structure. PLoS ONE 2011, 6, e26603. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Feiveson, A.H.; Krieger, S.; Kay Brinda, A.; von Scheven, G.; Bürkle, A.; Crucian, B.; Wu, H. Synergistic effects of weightlessness, isoproterenol, and radiation on DNA damage response and cytokine production in immune cells. Int. J. Mol. Sci. 2018, 19, 3689. [Google Scholar] [CrossRef]
- Canova, S.; Fiorasi, F.; Mognato, M.; Grifalconi, M.; Reddi, E.; Russo, A.; Celotti, L. “Modeled microgravity” affects cell response to ionizing radiation and increases genomic damage. Radiat. Res. 2005, 163, 191–199. [Google Scholar] [CrossRef]
- Girardi, C.; De Pittà, C.; Casara, S.; Sales, G.; Lanfranchi, G.; Celotti, L.; Mognato, M. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS ONE 2012, 7, e31293. [Google Scholar] [CrossRef]
- Mosesso, P.; Schuber, M.; Seibt, D.; Schmitz, C.; Fiore, M.; Schinoppi, A.; Penna, S.; Palitti, F. X-ray-induced chromosome aberrations in human lymphocytes in vitro are potentiated under simulated microgravity conditions (Clinostat). Phys. Med. 2001, 17, S264–S266. [Google Scholar]
- Ding, L.H.; Park, S.; Peyton, M.; Girard, L.; Xie, Y.; Minna, J.D.; Story, M.D. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to γ-rays and different elemental particles of high Z and energy. BMC Genom. 2013, 14, 372. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Dvorochkin, N.; Lee, C.; Alwood, J.S.; Yousuf, R.; Pianetta, P.; Globus, R.K.; Burns, B.P.; Almeida, E.A. Microgravity induces pelvic bone loss through osteoclastic activity, osteocytic osteolysis, and osteoblastic cell cycle inhibition by CDKN1a/p21. PLoS ONE 2013, 8, e61372. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Tian, H.; Zhang, J.; Qian, J.; Li, L.; Shi, L.; Zhao, Y. Spaceflight/microgravity inhibits the proliferation of hematopoietic stem cells by decreasing Kit-Ras/cAMP-CREB pathway networks as evidenced by RNA-Seq assays. FASEB J. 2019, 33, 5903–5913. [Google Scholar] [CrossRef] [PubMed]
- Ikenaga, M.; Hirayama, J.; Kato, T.; Kitao, H.; Han, Z.B.; Ishizaki, K.; Nishizawa, K.; Suzuki, F.; Cannon, T.F.; Fukui, K.; et al. Effect of space flight on the frequency of micronuclei and expression of stress-responsive proteins in cultured mammalian cells. J. Radiat. Res. 2002, 43, S141–S147. [Google Scholar] [CrossRef]
- Takahashi, A.; Suzuki, H.; Omori, K.; Seki, M.; Hashizume, T.; Shimazu, T.; Ishioka, N.; Ohnishi, T. Expression of p53-regulated proteins in human cultured lymphoblastoid TSCE5 and WTK1 cell lines during spaceflight. J. Radiat. Res. 2012, 53, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Ohnishi, K.; Takahashi, A.; Taniguchi, Y.; Sato, M.; Nakano, T.; Nagaoka, S. Detection of DNA damage induced by space radiation in Mir and space shuttle. J. Radiat. Res. 2002, 43, S133–S136. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Takahashi, A.; Nagamatsu, A.; Omori, K.; Suzuki, H.; Shimazu, T.; Ishioka, N. Detection of space radiation-induced double strand breaks as a track in cell nucleus. Biochem. Biophys. Res. Commun. 2009, 390, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Zhang, Y.; Wong, M.; Feiveson, A.; Gaza, R.; Stoffle, N.; Wang, H.; Wilson, B.; Rohde, L.; Stodieck, L.; et al. Detection of DNA damage by space radiation in human fibroblasts flown on the International Space Station. Life Sci. Space Res. 2017, 12, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef]
- Zhang, Y.; Moreno-Villanueva, M.; Krieger, S.; Ramesh, G.T.; Neelam, S.; Wu, H. Transcriptomics, NF-κB pathway, and their potential spaceflight-related health consequences. Int. J. Mol. Sci. 2017, 18, 1166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, T.; Wong, M.; Wang, X.; Stodieck, L.; Karouia, F.; Story, M.; Wu, H. Transient gene and microRNA expression profile changes of confluent human fibroblast cells in spaceflight. FASEB J. 2016, 30, 2211–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, T.G.; Lewis, F.C.; Goodwin, T.J.; Linnehan, R.M.; Wolf, D.A.; Hire, K.P.; Campbel, W.C.; Benes, E.; O’Reilly, K.C.; Globus, R.K.; et al. Gene expression in space. Nat. Med. 1999, 5, 359. [Google Scholar] [CrossRef]
- Harada-Sukeno, A.; Kohno, S.; Nakano, R.; Hirasaka, K.; Higashibata, A.; Yamazaki, T.; Ishioka, N.; Suzuki, H.; Shimazu, T.; Maita, A.; et al. Myo Lab: A JAXA cell biology experiment in “Kibo (JEM)” of the International Space Station. Biol. Sci. Space 2009, 23, 189–193. [Google Scholar] [CrossRef]
- Tanigawa, N.; Yano, S.; Higashibata, A.; Tsuchiya, Y.; Tanii, H.; Ando, N.; Kubota, K.; Nagase, M. Development and validation of a closed chamber for cell culture experiments in space. Biol. Sci. Space 2014, 28, 1–5. [Google Scholar] [CrossRef]
- Ohno, T.; Kanai, T.; Yamada, S.; Yusa, K.; Tashiro, M.; Shimada, H.; Torikai, K.; Yoshida, Y.; Kitada, Y.; Katoh, H.; et al. Carbon ion radio-therapy at the Gunma university heavy ion medical center: New facility set-up. Cancers 2011, 3, 4046–4060. [Google Scholar] [CrossRef] [PubMed]






| (a) Genes up-regulated by radiation and/or simulated μG | ||
| vs. ST | Total Genes | Cellular Pathways (Number of Genes) |
| ST-X3 | 315 | p53 signaling (7), FoxO signaling (5), Adrenergic signaling in cardiomyocytes (5) |
| ST-X24 | 523 | Neuroactive ligand–receptor interaction (13), Calcium signaling (11), cAMP signaling (11) |
| ST-C3 | 253 | Cytokine–cytokine receptor interaction (7), p53 signaling (6), Measles (5) |
| ST-C24 | 350 | p53 signaling (6), FoxO signaling (5) |
| RO-X3 | 204 | p53 signaling (4) |
| RO-X24 | 674 | Neuroactive ligand–receptor interaction (17), Calcium signaling (11), cAMP signaling (11) |
| RO-C3 | 211 | p53 signaling (7), Cytokine–cytokine receptor interaction (6) |
| RO-C24 | 339 | PI3K–Akt signaling (10), p53 signaling (6), ABC transporters (5) |
| RO | 140 | Morphine addiction (3) |
| (b) Genes down-regulated by radiation and/or simulated μG | ||
| vs. ST | Total Genes | Cellular Pathways (Number of Genes) |
| ST-X3 | 79 | Cell cycle (5), MicroRNAs in cancer (4), p53 signaling (3) |
| ST-X24 | 439 | Systemic lupus erythematosus (46), Alcoholism (46), Cell cycle (32) |
| ST-C3 | 198 | Cell cycle (5), Systemic lupus erythematosus (4) |
| ST-C24 | 663 | Systemic lupus erythematosus (55), Alcoholism (55), Cell cycle (36) |
| RO-X3 | 86 | Pathways in cancer (5) |
| RO-X24 | 507 | Alcoholism (46), Systemic lupus erythematosus (45), Cell cycle (33) |
| RO-C3 | 210 | Pathways in cancer (7) |
| RO-C24 | 702 | Systemic lupus erythematosus (55), Alcoholism (55), Cell cycle (35) |
| RO | 103 | * Calcium ion binding (6), * G-protein coupled purinergic nucleotide receptor activity (2), * Myosin binding (2) |
| Cell Cycle | Gene ID (Protein) | Ensembl | Function | Ref. |
|---|---|---|---|---|
| Suppression | ABL1 (c-Abl) | ENSG00000097007 | This gene encodes a protein tyrosine kinase. C-Abl protects p53 by antagonizing the inhibitory effect of Mdm2, an action that requires direct interplay between c-Abl and Mdm2. | [39] |
| CDKN1A (p21) | ENSG00000124762 | The encoded protein binds to and inhibits the activity of cyclin D1–CDK4/6 or cyclin B1–CDK1 complexes, and thus functions as a regulator of cell cycle progression at G1 and G2. | [40,41] | |
| Promotion | CCNB1 (Cyclin B1) | ENSG00000134057 | Activated cyclin B1 with CDK1 promotes several of the events of early mitosis. DNA damage leads to nuclear accumulation of inactive cyclin B1–CDK1 complexes by p21, and contributes to the establishment of permanent G2 arrest. | [40] |
| CCND1 (Cyclin D1) | ENSG00000110092 | This cyclin forms a complex with and functions as a regulatory subunit of CDK4/6, whose activity is required for cell cycle G1/S transition. DNA damage leads to nuclear accumulation of inactive cyclin D1–CDK4/6 complexes by p21, and contributes to the establishment of G1 arrest. | [41] | |
| KPNA2 (KPNA2) | ENSG00000182481 | KPNA2 expression accelerates cell cycle progression by up-regulating cyclin B and CDK1. | [42] | |
| MCM4 (MCM4) | ENSG00000104738 | MCM4, a subunit of a putative replicative helicase, is essential for the initiation of eukaryotic genome replication. MCM4 is one of the crucial targets of the DNA replication checkpoint system. | [43] | |
| MDM2 (MDM2) | ENSG00000135679 | MDM2 can promote tumor formation by targeting tumor suppressor p53 proteins for proteasomal degradation. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase. | [44] | |
| MKI67 (Ki-67) | ENSG00000148773 | Ki-67 is associated with and may be necessary for cellular proliferation. Ki-67 contributes to normal cell cycle progression. | [45] | |
| STMN1 (Stathmin 1) | ENSG00000117632 | Stathmin 1 is a ubiquitous cytosolic phosphor-protein. Stathmin is critically important not only for the formation of a normal mitotic spindle upon entry into mitosis but also for regulation of the function of the mitotic spindle in the later stages of mitosis and for the timely exit from mitosis. | [46] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, H.; Muratani, M.; Hidema, J.; Hada, M.; Fujiwara, K.; Souda, H.; Yoshida, Y.; Takahashi, A. Expression Profile of Cell Cycle-Related Genes in Human Fibroblasts Exposed Simultaneously to Radiation and Simulated Microgravity. Int. J. Mol. Sci. 2019, 20, 4791. https://doi.org/10.3390/ijms20194791
Ikeda H, Muratani M, Hidema J, Hada M, Fujiwara K, Souda H, Yoshida Y, Takahashi A. Expression Profile of Cell Cycle-Related Genes in Human Fibroblasts Exposed Simultaneously to Radiation and Simulated Microgravity. International Journal of Molecular Sciences. 2019; 20(19):4791. https://doi.org/10.3390/ijms20194791
Chicago/Turabian StyleIkeda, Hiroko, Masafumi Muratani, Jun Hidema, Megumi Hada, Keigi Fujiwara, Hikaru Souda, Yukari Yoshida, and Akihisa Takahashi. 2019. "Expression Profile of Cell Cycle-Related Genes in Human Fibroblasts Exposed Simultaneously to Radiation and Simulated Microgravity" International Journal of Molecular Sciences 20, no. 19: 4791. https://doi.org/10.3390/ijms20194791
APA StyleIkeda, H., Muratani, M., Hidema, J., Hada, M., Fujiwara, K., Souda, H., Yoshida, Y., & Takahashi, A. (2019). Expression Profile of Cell Cycle-Related Genes in Human Fibroblasts Exposed Simultaneously to Radiation and Simulated Microgravity. International Journal of Molecular Sciences, 20(19), 4791. https://doi.org/10.3390/ijms20194791