Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants
Abstract
:1. Introduction
2. CPK Enzymes and Related Kinases
3. CPK Family in Plants
3.1. CPK Distribution and Localization in Plants
3.2. CPK Domain Organization and Calcium Ion Signal Decryption
3.3. Functional Characterization of Plant CPKs
4. Role of CPKs in Abiotic Stress Tolerance
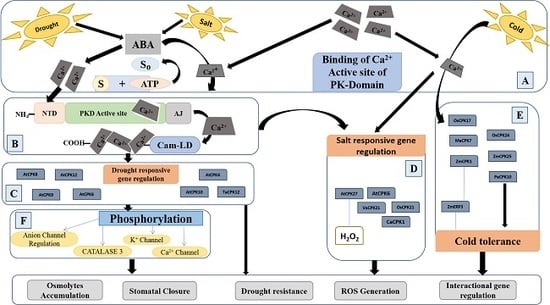
4.1. CPK-Mediated Drought Response Signaling
4.2. CPKs-Mediated Salt Response Signaling
4.3. CPK-Dependent Cold and Heat Stress Signaling
4.4. Role of CPKs in ROS Detoxification
5. Functional Interaction of CPKs with Other Kinases in Abiotic Stress Signaling
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AJ | Autoinhibitory junction |
| APX | Ascorbate peroxidase |
| AtABI1 | Arabidopsis thaliana ABA-insenstive-1 |
| AtCDPK/AtCPK | Arabidopsis thaliana-calcium dependent protein |
| AtCSD1 | Arabidopsis thaliana copper superoxide dismutase 1 |
| AtHSP101 and 70 | Arabidopsis thaliana heat shock protein 101 and 70 |
| AtProT1 | Arabidopsis thaliana proline transporter 1 |
| AtRBOHD | Arabidopsis thaliana respiratory burst oxidase protein D |
| AtRBOHF | Arabidopsis thaliana respiratory burst oxidase protein F |
| BaCDPK/BaCPK | Brassica napus calcium dependent protein kinase |
| BIK1 | Botrytis-induced kinase 1 |
| bZIP | Basic leucine zipper domain |
| Ca | Calcium |
| Ca2+ | Calcium ion |
| CaCDPK/CaCPK | Cicer arietinum calcium dependent protein kinase |
| CaMs | Calmodulins |
| CAT | Catalase |
| CAT3 | Catalase-3 |
| CBD | Calcium binding domain |
| CBF4 | C-repeat binding factor 4 |
| CBLs | Calcineurin β-like proteins |
| CCaMK | Calcium/Calmodulin-dependent protein kinase |
| CDPKs/CPKs | Calcium dependent protein kinases |
| CMLs | Calmodulin-like protein Kinase |
| CNGC18 | Cyclic nucleotide-gated ion channel 18 |
| Co-IP | Co-immunoprecipitation assay |
| COR | Cold regulator |
| CT | C-terminus |
| CTR | C-repeat |
| DRE | Dehydration elements |
| EF | Elongation Factor |
| ERD1 | Early responsive to dehydration stress 1 |
| ERF3 | Ethylene response factor 3 |
| FaCDPK/FaCPK | Fragaria x ananassa calcium dependent protein kinase |
| GA | Gibberellic acid |
| GA3ox1 | Gibberellin-3-betaoxigenase 1 |
| GOLS1 | Galactinol synthase 1 |
| GPX | Glutathione peroxidase |
| GR | Glutathione reductase |
| H2O2 | Hydrogen peroxide |
| HSF | Heat stress transcription factor |
| HSP | Heat shock protein |
| HvCDPK/HvCPK | Hordeum vulgare calcium dependent protein kinase |
| JA | Jasmonic acid |
| K+ | Potassium ion |
| LeCDPK/LeCPK | Solanum lycopersicum calcium dependent protein kinase |
| MaCDPK/MaCPK | Musa acuminate calcium dependent protein kinase |
| MPK5 | Mitogen-activated protein kinase 5 |
| MsCPK | Medicago sativa calcium dependent protein kinase |
| NaCDPK/NaCPK | Nicotiana attenuate calcium dependent protein kinase |
| NADPH | Nicotinamide Adenine Dinucleotide Phosphate Hydrogen |
| NHX | Sodium/Hydrogen exchanger |
| NST | NAC-transcription factors |
| NtCDPK/NtCPK | Nicotiana tabacum calcium dependent protein kinase |
| N-VD | N-terminus variable domain |
| OsCPK/OsCDPK | Oryza sativa calcium dependent protein kinase |
| OsGrx10 | Oryza sativa glutaredoxin 10 |
| OX | Overexpression |
| P5CS1 | Pyrroline-5-carboxylate synthetase 1 |
| PaCDPK/PaCPK | Phalaenopsis amabilis calcium dependent protein kinase |
| PeCDPK/PeCPK | Populus euphratica calcium dependent protein kinase |
| PEG | Polyethylene glycol |
| PEST | Proline, glutamine, serine and threonine |
| PgCDPK/PgCPK | Panax ginseng calcium dependent protein kinase |
| PIP | Plasma membrane intrinsic protein |
| PKD | Protein kinase domain |
| PnCDPK/PnCPK | Populus euphratica calcium dependent protein kinase |
| PRR | Pattern recognition receptor |
| qRT-PCR | Quantitative reverse transcription Polymerase chain reaction |
| RAB18 | Ras-associated binding protein 18 |
| RbohD | Respiratory burst oxidase homolog protein D |
| RD2A | Dehydration responsive protein 2A |
| RD29A | Dehydration responsive protein 29A |
| ROS | Reactive oxygen species |
| RT-PCR | Real-time PCR |
| SiCDPK/SiCPK | Setaria italic calcium dependent protein kinase |
| SOD | Superoxide dismutase |
| StCDPK/StCPK | Solanum tuberosum calcium dependent protein kinase |
| TaCDPK/TaCPK | Triticum aestivum calcium dependent protein kinase |
| T-DNA | Transfer DNA |
| VaCDPK/VaCPK | Vitis amurensis calcium dependent protein kinase |
| VaCPK/VaCDPK | Vitis amurenssis calcium dependent protein kinase |
| VfCPK/VfCDPK | Vicia faba calcium dependent protein kinase |
| VrCDPK/VrCPK | Vigna radiata calcium dependent protein kinase |
| ZmCDPK1/ZmCPK1 | Zea mays calcium dependent protein kinase 1 |
| ZoCDPK/ZoCPK | Zingiber officinale calcium dependent protein kinase |
References
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, H.; Wei, X.; Yang, L.; Yang, B.; Zhang, L.; Li, J.; Jiang, Y.-Q. Functional characterization of calcium-dependent protein kinase (CPK) 2 gene from oilseed rape (Brassica napus L.) in regulating reactive oxygen species signaling and cell death control. Gene 2018, 651, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wang, X.; Leseberg, C.H.; Jia, J.; Mao, L. Biotic and abiotic stress responses through calcium-dependent protein kinase (CDPK) signaling in wheat (Triticum aestivum L.). Plant Sig. Behav. 2008, 3, 654–656. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kikuchi, S.; Ohsugi, R. CDPK-mediated abiotic stress signaling. Plant Sig. Behav. 2012, 7, 817–821. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.-C.; Chen, J.; Miao, C.; Song, C.-P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Jiang, M.; Zhang, A.; Lu, J. Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 2005, 223, 57. [Google Scholar] [CrossRef]
- Baba, A.I.; Rigó, G.; Andrási, N.; Tietz, O.; Palme, K.; Szabados, L.; Cséplő, Á. Striving Towards Abiotic Stresses: Role of the Plant CDPK Superfamily Members; Springer: Berlin, Germany, 2019; pp. 99–105. [Google Scholar]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef]
- Ray, S.D. Decrypting Calcium Signaling in Plants: The Kinase Way; Springer: Berlin, Germany, 2015; pp. 119–174. [Google Scholar]
- Mohanta, T.K.; Yadav, D.; Khan, A.L.; Hashem, A.; Abd Allah, E.F.; Al-Harrasi, A. Molecular Players of EF-hand Containing Calcium Signaling Event in Plants. Int. J. Mol. Sci. 2019, 20, 1476. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin B–like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14, S389–S400. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Kudla, J. Calcium decoding mechanisms in plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.; Trewavas, A. Activation of a pea membrane protein kinase by calcium ions. Planta 1984, 161, 409–417. [Google Scholar] [CrossRef]
- Harmon, A.C.; Gribskov, M.; Harper, J.F. CDPKs–A kinase for every Ca2+ signal? Trends Plant Sci. 2000, 5, 154–159. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef]
- Rudd, J.J.; Franklin-Tong, V.E. Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol. 2001, 151, 7–33. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed]
- Initiative, A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Agarwal, P.; Arora, R.; Kapoor, S.; Tyagi, A.K. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genom. 2007, 278, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-L.; Zhu, Y.-F.; Tan, X.-M.; Wang, X.; Wei, B.; Guo, H.-Z.; Zhang, Z.-L.; Chen, X.-B.; Zhao, G.-Y.; Kong, X.-Y. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xia, E.-H.; Gao, L.-Z. Genome-wide analysis of WRKY family of transcription factors in common bean, Phaseolus vulgaris: Chromosomal localization, structure, evolution and expression divergence. Plant Gene 2016, 5, 22–30. [Google Scholar] [CrossRef]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS ONE 2014, 9, e98189. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.-T.; Zhao, F.-L.; Hu, Y.; Gao, Y.-R.; Ma, Y.-F.; Zheng, Y.; Wang, Y.-J.; Wen, Y.-Q. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 2015, 15, 164. [Google Scholar] [CrossRef]
- Hu, W.; Hou, X.; Xia, Z.; Yan, Y.; Wei, Y.; Wang, L.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava. Mol. Genet. Genom. 2016, 291, 241–253. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Q.; Chen, Q.; Yin, X.; Qian, M.; Sun, X.; Yang, Y. Genome-wide survey indicates diverse physiological roles of the barley (Hordeum vulgare L.) calcium-dependent protein kinase genes. Sci. Rep. 2017, 7, 5306. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, C.; Yang, X.; Chen, H.; Yang, Y.; Mo, Y.; Li, H.; Zhang, Y.; Ma, J.; Yang, J. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS ONE 2017, 12, e0176352. [Google Scholar] [CrossRef]
- Weckwerth, P.; Ehlert, B.; Romeis, T. Zm CPK 1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015, 38, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Zuo, R.; Hu, R.; Chai, G.; Xu, M.; Qi, G.; Kong, Y.; Zhou, G. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol. Biol. Rep. 2013, 40, 2645–2662. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Wang, L.; Xie, W.; Wan, B.; Li, X.; Lin, Y. Expression profile of calcium-dependent protein kinase (CDPKs) genes during the whole lifespan and under phytohormone treatment conditions in rice (Oryza sativa L. ssp. indica). Plant Mol. Biol. 2009, 70, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Matschi, S.; Shorinola, O.; Rovenich, H.; Matei, A.; Segonzac, C.; Malinovsky, F.G.; Rathjen, J.P.; MacLean, D.; Romeis, T. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 2014, 16, 605–615. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Mohanta, N.; Mohanta, Y.K.; Bae, H. Genome-wide identification of calcium dependent protein kinase gene family in plant lineage shows presence of novel DxD and DEL motifs in EF-hand domain. Front. Plant Sci. 2015, 6, 1146. [Google Scholar] [CrossRef]
- Li, M.; Hu, W.; Ren, L.; Jia, C.; Liu, J.; Miao, H.; Guo, A.; Xu, B.; Jin, Z. Identification, Expression, and Interaction Network Analyses of the CDPK Gene Family Reveal Their Involvement in the Development, Ripening, and Abiotic Stress Response in Banana. Biochem. Genet. 2019, 1–23. [Google Scholar] [CrossRef]
- Fedorowicz-Strońska, O.; Koczyk, G.; Kaczmarek, M.; Krajewski, P.; Sadowski, J. Genome-wide identification, characterisation and expression profiles of calcium-dependent protein kinase genes in barley (Hordeum vulgare L.). J. App. Genet. 2017, 58, 11–22. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.-Z.; Zhang, Y.; Deng, M.; Niu, F.; Yang, B.; Wang, X.; Wang, B.; Liang, W.; Deyholos, M.K. Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.). BMC Genom. 2014, 15, 211. [Google Scholar] [CrossRef]
- Tong, X.; Cao, A.; Wang, F.; Chen, X.; Xie, S.; Shen, H.; Jin, X.; Li, H. Calcium-Dependent Protein Kinase Genes in Glycyrrhiza Uralensis Appear to be Involved in Promoting the Biosynthesis of Glycyrrhizic Acid and Flavonoids under Salt Stress. Molecules 2019, 24, 1837. [Google Scholar] [CrossRef]
- Gao, W.; Xu, F.-C.; Guo, D.-D.; Zhao, J.-R.; Liu, J.; Guo, Y.-W.; Singh, P.K.; Ma, X.-N.; Long, L.; Botella, J.R. Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 2018, 18, 15. [Google Scholar] [CrossRef]
- Xu, X.; Liu, M.; Lu, L.; He, M.; Qu, W.; Xu, Q.; Qi, X.; Chen, X. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.M.; Chandra, M.; Shankhdhar, S.C.; Kumar, A. Transcriptome wide identification and validation of calcium sensor gene family in the developing spikes of finger millet genotypes for elucidating its role in grain calcium accumulation. PLoS ONE 2014, 9, e103963. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-F.; Zhao, W.-Y.; Fu, J.-D.; Liu, Y.-W.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; Xi, Y.-J. Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fasoli, M.; Tornielli, G.B.; Dal Santo, S.; Pezzotti, M.; Zhang, L.; Cai, B.; Cheng, Z.-M. The evolutionary history and diverse physiological roles of the grapevine calcium-dependent protein kinase gene family. PLoS ONE 2013, 8, e80818. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S. Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis. J. Plant Physiol. 2013, 170, 1491–1500. [Google Scholar] [CrossRef]
- Ma, P.; Liu, J.; Yang, X.; Ma, R. Genome-wide identification of the maize calcium-dependent protein kinase gene family. App. Biochem. Biotechnol. 2013, 169, 2111–2125. [Google Scholar] [CrossRef]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef]
- He, S. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum. Front. Plant Sci. 2015, 6, 737. [Google Scholar]
- Wankhede, D.P.; Kumari, M.; Richa, T.; Aravind, J.; Rajkumar, S. Genome wide identification and characterization of Calcium Dependent Protein Kinase gene family in Cajanus cajan. J. Environ. Biol. 2017, 38, 167. [Google Scholar] [CrossRef]
- Gromadka, R.; Cieśla, J.; Olszak, K.; Szczegielniak, J.; Muszyńska, G.; Polkowska-Kowalczyk, L. Genome-wide analysis and expression profiling of calcium-dependent protein kinases in potato (Solanum tuberosum). Plant Growth Reg. 2018, 84, 303–315. [Google Scholar] [CrossRef]
- Asano, T.; Tanaka, N.; Yang, G.; Hayashi, N.; Komatsu, S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005, 46, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Wan, B.; Lin, Y.; Mou, T. Expression of rice Ca2+-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett. 2007, 581, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.H.; Yang, M.; Sui, J.L.; Qi, J.Y.; Fang, Y.J.; Hu, S.N.; Tang, C.R. The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis—Comparison with five other plant species in structure, evolution, and expression. FEBS Open Biol. 2017, 7, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, F. Calcium-Dependent Protein Kinase Regulates Soybean Serine Acetyltransferase in Response to Oxidative Stress. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2002. [Google Scholar]
- Hettenhausen, C.; Sun, G.; He, Y.; Zhuang, H.; Sun, T.; Qi, J.; Wu, J. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 2016, 6, 18973. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Che, Z.; Zeng, X.; Zhou, X.; Sitoe, H.M.; Wang, H.; Yu, D. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integ. Genom. 2016, 16, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.-S.; Liu, G.-S.; Sun, Y.-H.; Jia, C. Cloning and expression of calcium-dependent protein kinase (CDPK) gene family in common tobacco (Nicotiana tabacum). Agric. Sci. China 2009, 8, 1448–1457. [Google Scholar] [CrossRef]
- Wang, J.-P.; Xu, Y.-P.; Munyampundu, J.-P.; Liu, T.-Y.; Cai, X.-Z. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef]
- Hu, Z.; Lv, X.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y. Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 2016, 7, 469. [Google Scholar] [CrossRef]
- Podell, S.; Gribskov, M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genom. 2004, 5, 37. [Google Scholar] [CrossRef]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta BBA Mol. Cell Res. 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [Green Version]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Himschoot, E.; Beeckman, T.; Friml, J.; Vanneste, S. Calcium is an organizer of cell polarity in plants. Biochim. Biophys. Acta BBA Mol. Cell Res. 2015, 1853, 2168–2172. [Google Scholar] [CrossRef] [PubMed]
- Harmon, A.C.; Gribskov, M.; Gubrium, E.; Harper, J.F. The CDPK superfamily of protein kinases. New Phytol. 2001, 151, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Grabarek, Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J. Mol. Biol. 2006, 359, 509–525. [Google Scholar] [CrossRef]
- Klimecka, M.; Muszynska, G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. Eng. Edi. 2007, 54, 219. [Google Scholar]
- Wernimont, A.K.; Artz, J.D.; Finerty, P., Jr.; Lin, Y.-H.; Amani, M.; Allali-Hassani, A.; Senisterra, G.; Vedadi, M.; Tempel, W.; Mackenzie, F. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 2010, 17, 596. [Google Scholar] [CrossRef]
- Parvathy, S.T. Versatile roles of ubiquitous calcium-dependent protein kinases (CDPKs) in plants. Indian Soc. Oilseeds Res. 2018, 35, 1–11. [Google Scholar]
- Wei, S.; Hu, W.; Deng, X.; Zhang, Y.; Liu, X.; Zhao, X.; Luo, Q.; Jin, Z.; Li, Y.; Zhou, S. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014, 14, 133. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Deng, C.; Deng, S.; Li, N.; Zhao, C.; Zhao, R.; Liang, S.; Chen, S. The Arabidopsis Ca2+-Dependent Protein Kinase CPK12 Is Involved in Plant Response to Salt Stress. Int. J. Mol. Sci. 2018, 19, 4062. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. The calcium-dependent protein kinase gene VaCPK29 is involved in grapevine responses to heat and osmotic stresses. Plant Growth Reg. 2017, 82, 79–89. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-H.; Liu, H.-P.; Li, C.-L. Calcium-dependent protein kinase CPK9 negatively functions in stomatal abscisic acid signaling by regulating ion channel activity in Arabidopsis. Plant Mol. Biol. 2019, 99, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Coca, M.; San Segundo, B. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 2010, 63, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Fantino, E.; Segretin, M.E.; Santin, F.; Mirkin, F.G.; Ulloa, R.M. Analysis of the potato calcium-dependent protein kinase family and characterization of StCDPK7, a member induced upon infection with Phytophthora infestans. Plant Cell Rep. 2017, 36, 1137–1157. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.-P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.; Park, J.M.; Oh, S.-K.; Joung, Y.H.; Lee, S.; Choi, D. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta 2004, 220, 286–295. [Google Scholar] [CrossRef]
- Yang, D.-H.; Hettenhausen, C.; Baldwin, I.T.; Wu, J. Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound-and herbivory-induced jasmonic acid accumulations. Plant Physiol. 2012, 159, 1591–1607. [Google Scholar] [CrossRef]
- Matschi, S.; Werner, S.; Schulze, W.X.; Legen, J.; Hilger, H.H.; Romeis, T. Function of calcium-dependent protein kinase CPK 28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 2013, 73, 883–896. [Google Scholar] [CrossRef]
- Jin, Y.; Ye, N.; Zhu, F.; Li, H.; Wang, J.; Jiang, L.; Zhang, J. Calcium-dependent protein kinase CPK 28 targets the methionine adenosyltransferases for degradation by the 26S proteasome and affects ethylene biosynthesis and lignin deposition in Arabidopsis. Plant J. 2017, 90, 304–318. [Google Scholar] [CrossRef]
- Zhou, L.; Lan, W.; Jiang, Y.; Fang, W.; Luan, S. A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant 2014, 7, 369–376. [Google Scholar] [CrossRef]
- Zhu, J.-H.; Chen, X.; Chang, W.-J.; Tian, W.-M.; Zhang, Z.-L. Molecular characterization of HbCDPK1, an ethephon-induced calcium-dependent protein kinase gene of Hevea brasiliensis. Biosci. Biotechnol. Biochem. 2010, 74, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Turlenko, A.V.; Zhuravlev, Y.N. Structure and expression profiling of a novel calcium-dependent protein kinase gene PgCDPK1a in roots, leaves, and cell cultures of Panax ginseng. Plant Cell Tissue Organ Cult. 2010, 103, 197–204. [Google Scholar] [CrossRef]
- Tsai, T.-M.; Chen, Y.-R.; Kao, T.-W.; Tsay, W.-S.; Wu, C.-P.; Huang, D.-D.; Chen, W.-H.; Chang, C.-C.; Huang, H.-J. PaCDPK1, a gene encoding calcium-dependent protein kinase from orchid, Phalaenopsis amabilis, is induced by cold, wounding, and pathogen challenge. Plant Cell Rep. 2007, 26, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, D.; Wang, L.; Pan, J.; Liu, Y.; Kong, X.; Zhou, Y.; Li, D. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 71, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.; Dubrovina, A.; Shumakova, O.; Karetin, Y.; Manyakhin, A. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013, 32, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Aleynova, O.; Dubrovina, A.; Kiselev, K. Activation of stilbene synthesis in cell cultures of Vitis amurensis by calcium-dependent protein kinases VaCPK1 and VaCPK26. Plant Cell Tissue Organ Cult. 2017, 130, 141–152. [Google Scholar] [CrossRef]
- Pawełek, A.; Szmidt-Jaworska, A.; Świeżawska, B.; Jaworski, K. Genomic structure and promoter characterization of the CDPK kinase gene expressed during seed formation in Pharbitis nil. J. Plant Physiol. 2015, 189, 87–96. [Google Scholar] [CrossRef]
- Martínez-Noël, G.; Nagaraj, V.J.; Caló, G.; Wiemken, A.; Pontis, H.G. Sucrose regulated expression of a Ca2+-dependent protein kinase (TaCDPK1) gene in excised leaves of wheat. Plant Physiol. Biochem. 2007, 45, 410–419. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium dependent protein kinase, a versatile player in plant stress management and development. Critic. Rev. Plant Sci. 2017, 36, 336–352. [Google Scholar] [CrossRef]
- Monroy, A.F.; Dhindsa, R.S. Low-temperature signal transduction: Induction of cold acclimation-specific genes of alfalfa by calcium at 25 °C. Plant Cell 1995, 7, 321–331. [Google Scholar] [PubMed]
- Fallon, K.M.; Shacklock, P.S.; Trewavas, A.J. Detection in vivo of very rapid red light-induced calcium-sensitive protein phosphorylation in etiolated wheat (Triticum aestivum) leaf protoplasts. Plant Physiol. 1993, 101, 1039–1045. [Google Scholar] [CrossRef] [PubMed]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 2000, 23, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.-J.; Li, X.-D.; Ratnasekera, D.; Wang, C.; Liu, W.-X.; Song, L.-F.; Zhang, W.-Z.; Wu, W.-H. Arabidopsis Calcium-Dependent Protein Kinase8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef]
- Zhu, S.-Y.; Yu, X.-C.; Wang, X.-J.; Zhao, R.; Li, Y.; Fan, R.-C.; Shang, Y.; Du, S.-Y.; Wang, X.-F.; Wu, F.-Q. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef]
- Takahashi, K.; Isobe, M.; Muto, S. An increase in cytosolic calcium ion concentration precedes hypoosmotic shock-induced activation of protein kinases in tobacco suspension culture cells. FEBS Lett. 1997, 401, 202–206. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Yoshioka, M.; Asai, S.; Nomura, H.; Kuchimura, K.; Mori, H.; Doke, N.; Yoshioka, H. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012, 196, 223–237. [Google Scholar] [CrossRef]
- Perochon, A.; Aldon, D.; Galaud, J.-P.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef]
- Szczegielniak, J.; Borkiewicz, L.; Szurmak, B.; Lewandowska-Gnatowska, E.; Statkiewicz, M.; Klimecka, M.; Cieśla, J.; Muszyńska, G. Maize calcium-dependent protein kinase (ZmCPK11): Local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol. Plant. 2012, 146, 1–14. [Google Scholar] [CrossRef]
- Ding, Y.; Cao, J.; Ni, L.; Zhu, Y.; Zhang, A.; Tan, M.; Jiang, M. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J. Exp. Bot. 2012, 64, 871–884. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.-J.; Wei, F.-J.; Wang, C.; Wu, J.-J.; Ratnasekera, D.; Liu, W.-X.; Wu, W.-H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid-and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 2015, 185, 1–12. [Google Scholar] [CrossRef]
- Liu, G.; Chen, J.; Wang, X. VfCPK1, a gene encoding calcium-dependent protein kinase from Vicia faba, is induced by drought and abscisic acid. Plant Cell Environ. 2006, 29, 2091–2099. [Google Scholar] [CrossRef]
- Ratnasekera, D. A calcium dependent protein kinase involves H2O2 mediated guard cell signaling in Aarabidopsis. Trop. Agri. Res. Exten. 2015, 16, 7–14. [Google Scholar] [CrossRef]
- Syam Prakash, S.R.; Jayabaskaran, C. Heterologous expression and biochemical characterization of two calcium-dependent protein kinase isoforms CaCPK1 and CaCPK2 from chickpea. J. Plant Physiol. 2006, 163, 1083–1093. [Google Scholar] [CrossRef]
- Ho, S.-L.; Huang, L.-F.; Lu, C.-A.; He, S.-L.; Wang, C.-C.; Yu, S.-P.; Chen, J.; Yu, S.-M. Sugar starvation-and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol. Biol. 2013, 81, 347–361. [Google Scholar] [CrossRef]
- Geng, S.; Zhao, Y.; Tang, L.; Zhang, R.; Sun, M.; Guo, H.; Kong, X.; Li, A.; Mao, L. Molecular evolution of two duplicated CDPK genes CPK7 and CPK12 in grass species: A case study in wheat (Triticum aestivum L.). Gene 2011, 475, 94–103. [Google Scholar] [CrossRef]
- Vivek, P.J.; Tuteja, N.; Soniya, E.V. CDPK1 from ginger promotes salinity and drought stress tolerance without yield penalty by improving growth and photosynthesis in Nicotiana tabacum. PLoS ONE 2013, 8, e76392. [Google Scholar] [CrossRef]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B. Specific Calcium and Abscisic Acid Regulation of Anion Channels in Arabidopsis Guard Cells. Ph.D. Thesis, Eberhard Karls University Tübingen, Tübingen, Germany, 2014. [Google Scholar]
- Baba, A.; Rigó, G.; Ayaydin, F.; Rehman, A.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.-F.; Andreoli, S.; Tiriac, H.; Alonso, J.M.; Harper, J.F.; Ecker, J.R. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion-and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [PubMed]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmann-Rodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, Y.-S.; Peng, R.-H.; Xiong, A.-S.; Zhu, B.; Jin, X.-F.; Gao, F.; Fu, X.-Y.; Hou, X.-L.; Yao, Q.-H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Milla, M.A.R.; Townsend, J.; Chang, F.; Cushman, J.C. The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol. Biol. 2006, 61, 13–30. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.L.; Mei, C.; Wang, X.J.; Yan, L.; Liu, R.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. The Arabidopsis Ca2+-dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011, 192, 61–73. [Google Scholar] [CrossRef]
- Huang, S.-J.; Chang, C.-L.; Wang, P.-H.; Tsai, M.-C.; Hsu, P.-H.; Chang, I.-F. A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 4343–4360. [Google Scholar] [CrossRef]
- Franz, S.; Ehlert, B.; Liese, A.; Kurth, J.; Cazalé, A.-C.; Romeis, T. Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 2011, 4, 83–96. [Google Scholar] [CrossRef]
- Ma, S.-Y.; Wu, W.-H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.; Zhao, N.; Jing, X.; Shen, X.; Chen, S. The Arabidopsis Ca2+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 2015, 563, 203–214. [Google Scholar] [CrossRef]
- Choi, H.-I.; Park, H.-J.; Park, J.H.; Kim, S.; Im, M.-Y.; Seo, H.-H.; Kim, Y.-W.; Hwang, I.; Kim, S.Y. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005, 139, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, N.; Sasabe, M.; Endo, M.; Machida, Y.; Araki, T. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 2015, 5, 8341. [Google Scholar] [CrossRef] [PubMed]
- Llop-Tous, I.; Domínguez-Puigjaner, E.; Vendrell, M. Characterization of a strawberry cDNA clone homologous to calcium-dependent protein kinases that is expressed during fruit ripening and affected by low temperature. J. Exp. Bot. 2002, 53, 2283–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davletova, S.; Mészáros, T.; Miskolczi, P.; Oberschall, A.; Török, K.; Magyar, Z.; Dudits, D.; Deák, M. Auxin and heat shock activation of a novel member of the calmodulin like domain protein kinase gene family in cultured alfalfa cells. J. Exp. Bot. 2001, 52, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S.; Hu, K.; Yang, J.; Xin, X.; Zhou, W.; Fan, J.; Cui, F.; Mou, B.; Zhang, S. The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol. 2018, 176, 1835–1849. [Google Scholar] [CrossRef]
- Kang, C.H.; Moon, B.C.; Park, H.C.; Koo, S.C.; Chi, Y.H.; Cheong, Y.H.; Yoon, B.-D.; Lee, S.Y.; Kim, C.Y. Rice small C2-domain proteins are phosphorylated by calcium-dependent protein kinase. Mol. Cells 2013, 35, 381–387. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Yu, X.; An, C. Overexpression of constitutively active OsCPK10 increases Arabidopsis resistance against Pseudomonas syringae pv. tomato and rice resistance against Magnaporthe grisea. Plant Physiol. Biochem. 2013, 73, 202–210. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, Y.; Bi, Z.; Liu, Q.; Xu, T.; Yu, N.; Cao, Y.; Zhu, A.; Wu, W.; Zhan, X. Impaired function of the calcium-dependent protein kinase, OsCPK12, leads to early senescence in Rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 52. [Google Scholar] [CrossRef]
- Abbasi, F.; Onodera, H.; Toki, S.; Tanaka, H.; Komatsu, S. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 2004, 55, 541–552. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef]
- Asano, T.; Hakata, M.; Nakamura, H.; Aoki, N.; Komatsu, S.; Ichikawa, H.; Hirochika, H.; Ohsugi, R. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 2011, 75, 179–191. [Google Scholar] [CrossRef]
- Manimaran, P.; Mangrauthia, S.K.; Sundaram, R.; Balachandran, S. Constitutive expression and silencing of a novel seed specific calcium dependent protein kinase gene in rice reveals its role in grain filling. J. Plant Physiol. 2015, 174, 41–48. [Google Scholar] [CrossRef]
- Ishida, S.; Yuasa, T.; Nakata, M.; Takahashi, Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor repression of shoot growth in response to gibberellins. Plant Cell 2008, 20, 3273–3288. [Google Scholar] [CrossRef]
- Romeis, T.; Ludwig, A.A.; Martin, R.; Jones, J.D. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001, 20, 5556–5567. [Google Scholar] [CrossRef] [Green Version]
- Botella, J.R.; Arteca, J.M.; Somodevilla, M.; Arteca, R.N. Calcium-dependent protein kinase gene expression in response to physical and chemical stimuli in mungbean (Vigna radiata). Plant Mol. Biol. 1996, 30, 1129–1137. [Google Scholar] [CrossRef]
- Chang, W.-J.; Su, H.-S.; Li, W.-J.; Zhang, Z.-L. Expression profiling of a novel calcium-dependent protein kinase gene, LeCPK2, from tomato (Solanum lycopersicum) under heat and pathogen-related hormones. Biosci. Biotechnol. Biochem. 2009, 73, 24–27. [Google Scholar] [CrossRef]
- Santin, F.; Bhogale, S.; Fantino, E.; Grandellis, C.; Banerjee, A.K.; Ulloa, R.M. Solanum tuberosum StCDPK1 is regulated by miR390 at the posttranscriptional level and phosphorylates the auxin efflux carrier StPIN4 in vitro, a potential downstream target in potato development. Physiol. Plant. 2017, 159, 244–261. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Doke, N.; Yoshioka, H. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Sun, K.; Chen, X.; Zhou, Q.; Li, H.; Zhang, X.; Li, X. Involvement of CsCDPK20 and CsCDPK26 in Regulation of Thermotolerance in Tea Plant (Camellia sinensis). Plant Mol. Biol. Rep. 2018, 36, 176–187. [Google Scholar] [CrossRef]
- Freymark, G.; Diehl, T.; Miklis, M.; Romeis, T.; Panstruga, R. Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol. Plant Microbe Interact. 2007, 20, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gong, J.; Su, X.; Li, L.; Pang, X.; Zhang, Z. MaCDPK7, a calcium-dependent protein kinase gene from banana is involved in fruit ripening and temperature stress responses. J. Hort. Sci. Biotechnol. 2017, 92, 240–250. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Jia, C.; Xu, B.; Jin, Z. Molecular cloning and expression analysis of eight calcium-dependent protein kinase (CDPK) genes from banana (Musa acuminata L. AAA group, cv. Cavendish). S. Afr. J. Bot. 2016, 104, 134–141. [Google Scholar] [CrossRef]
- Dubrovina, A.; Kiselev, K. The Role of Calcium-Dependent Protein Kinase Genes VaCPK1 and VaCPK26 in the Response of Vitis amurensis (in vitro) and Arabidopsis thaliana (in vivo) to Abiotic Stresses. Russian J. Genet. 2019, 55, 319–329. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. VaCPK21, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., is involved in grape response to salt stress. Plant Cell. Tissue Organ Cult. 2016, 124, 137–150. [Google Scholar] [CrossRef]
- Chen, J.; Xue, B.; Xia, X.; Yin, W. A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Biophys. Res. Commun. 2013, 441, 630–636. [Google Scholar] [CrossRef]
- Ning, M.; Tang, F.; Zhang, Q.; Zhao, X.; Yang, L.; Cai, W.; Shan, C. Effects of Penicillium infection on the expression and activity of CDPK2 in postharvest Hami melon treated with calcium chloride. Physiol. Mol. Plant Pathol. 2019, 106, 175–181. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef]
- Dubrovina, A.; Aleynova, O.; Manyakhin, A.; Kiselev, K. The Role of Calcium-Dependent Protein Kinase Genes CPK16, CPK25, CPK30, and CPK32 in Stilbene Biosynthesis and the Stress Resistance of Grapevine Vitis amurensis Rupr. App. Biochem. Microbiol. 2018, 54, 410–417. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.-C.; Lu, Y.-T. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J. Plant Biol. 2013, 56, 306–314. [Google Scholar] [CrossRef]


| Sr. # | Common Name | Botanical Name | No. of CPKs | Genome Size (Mb) | Reference |
|---|---|---|---|---|---|
| 1 | Algae | Volvox carteri | 6 | 131.2 | [37] |
| 2 | Apple | Malus domestica | 28 | 881.3 | [37] |
| 3 | Arabidopsis | Arabidopsis thaliana | 34 | 135 | [20] |
| 4 | Banana | Musa acuminata | 44 | 523 | [38] |
| 5 | Barley | Hordeum vulgare | 27 | 667 | [39] |
| 6 | Barley | Hordeum vulgare | 28 | 667 | [31] |
| 7 | Barrel clover | Medicago truncatula | 11 | 360 | [37] |
| 8 | Black cottonwood | Populus trichocarpa | 28 | 422.9 | [37] |
| 9 | Poplar | Populus trichocarpa | 30 | 500 | [34] |
| 10 | Butcher | Micromonas pusilla | 22 | 2 | [13,37] |
| 11 | Cacao tree | Theobroma cacao | 17 | 346 | [13,37] |
| 12 | Canola | Brassica napus | 25 | 1130 | [40] |
| 13 | Cassava | Manihot esculenta | 26 | 532.5 | [30] |
| 14 | Caster bean | Ricinus communis | 15 | 400 | [37] |
| 15 | Castor bean | Ricinus communis | 15 | 400 | [13,37] |
| 16 | Chinese liquorice | Glycyrrhiza uralensis | 23 | 379 | [41] |
| 17 | Chlamydomonas | Chlamydomonas reinhardtii | 14 | 111.1 | [13,37] |
| 18 | Clementine | Citrus clememtina | 26 | 301.4 | [37] |
| 19 | Cocoa tree | Theobroma cacao | 17 | 346 | [37] |
| 20 | Columbine | Aquilegia coerulea | 16 | 306.5 | [13,37] |
| 21 | Cotton | Gossypium raimondii | 41 | 880 | [28] |
| 22 | Cotton | Gossypium hirsutum | 98 | 2250–2430 | [42] |
| 23 | Cucumber | Cucumis sativus | 19 | 323.99 | [43] |
| 24 | Cucumber | Cucumis sativus | 18 | 203 | [37] |
| 25 | Finger Millet | Eleusine coracana | 4 | 1593 | [44] |
| 26 | Flax | Linum usitatissimum | 47 | 318.3 | [37] |
| 27 | Flooded gum | Eucalyptus grandis | 22 | 691 | [37] |
| 28 | Foxtail Millet | Setaria italic | 27 | 405.7 | [37] |
| 29 | Foxtail Millet | Setaria italic | 29 | 405.7 | [45] |
| 30 | Foxtail millet | Setaria italica | 27 | 405.7 | [13,37] |
| 31 | Grape | Vitis vinifera | 19 | 500 | [29] |
| 32 | Grapevine | Vitis amurensis | 17 | 500 | [46] |
| 33 | Grapevine | Vitis amurensis | 13 | 500 | [47] |
| 34 | Green algae | Coccomyxa subellipsoidea | 2 | 49 | [13,37] |
| 35 | Green algae | Ostreococcus lucimarinus | 3 | 13.2 | [13,37] |
| 36 | Green bean | Phaseolus vulgaris | 25 | 521.1 | [37] |
| 37 | Linseed | Linum usitatissimum | 47 | 318.3 | [13,37] |
| 38 | Maize | Zea mays | 35 | 2500 | [48] |
| 39 | Maize | Zea mays | 40 | 2500 | [49] |
| 40 | Maize | Zea mays | 47 | 2500 | [37] |
| 41 | Melon | Cucumis melo | 18 | 375 | [32] |
| 42 | Monkey flower | Mimulus guttatus | 25 | 321.7 | [37] |
| 43 | Mustard | Brassica rapa | 49 | 283.8 | [37] |
| 44 | Norway spruce | Picea abies | 11 | 1960 | [37] |
| 45 | Oilseed rape | Brassica rapa | 49 | 283.8 | [13,37] |
| 46 | Orange | Citrus sinensis | 24 | 319 | [13,37] |
| 47 | Papaya | Carica papaya | 15 | 135 | [13,37] |
| 48 | Papaya | Carica papaya | 15 | 135 | [37] |
| 49 | Peach | Prunus persica | 17 | 227.3 | [37] |
| 50 | Pepper | Capsicum annuum | 31 | 407.5 | [50] |
| 51 | Pigeon Pea | Cajanus cajan | 23 | 852 | [51] |
| 52 | Potato | Solanum tubersum | 21 | 800 | [37] |
| 53 | Potato | Solanum tuberosum | 23 | 800 | [52] |
| 54 | Purple false brome | Brachypodium distachyon | 27 | 272 | [37] |
| 55 | Purple false brome | Brachipodium distachyon | 27 | 272 | [37] |
| 56 | Red Shepherd’s Purse | Capsella rubella | 32 | 134.8 | [37] |
| 57 | Rice | Oryza sativa | 29 | 430 | [53] |
| 58 | Rice | Oryza sativa | 22 | 430 | [54] |
| 59 | Rice | Oryza sativa | 30 | 372 | [37] |
| 60 | Rubber tree | Hevea brasiliensis | 30 | 1332 | [55] |
| 61 | Salt cress | Thellungiella halophile | 31 | 238.5 | [13,37] |
| 62 | Shepherd’s Purse | Capsella rubella | 32 | 134.8 | [37] |
| 63 | Sorghum | Sorghum bicolor | 28 | 697.5 | [37] |
| 64 | Soybean | Glycine max | 39 | 1115 | [56] |
| 65 | Soybean | Glycine max | 50 | 1115 | [57] |
| 66 | Soybean | Glycine max | 39 | 1115 | [58] |
| 67 | Soybean | Glycine max | 41 | 978 | [13,37] |
| 68 | Spikemosses | Selaginella moellendorffii | 11 | 212.5 | [13,37] |
| 69 | Spreading earthmoss | Physcomitrella patens | 25 | 480 | [13,37] |
| 70 | Sweet orange | Citrus sinensis | 24 | 319 | [37] |
| 71 | Switchgrass | Panicum virgatum | 53 | 1358 | [37] |
| 72 | Tobacco | Nicotiana tabacum | 15 | 323.75 | [59] |
| 73 | Tomato | Solanum lycopersicum | 29 | 900 | [60] |
| 74 | Tomato | Solanum lycopersicum | 28 | 900 | [37] |
| 75 | Tomato | Solanum lycopersicum | 29 | 900 | [61] |
| 76 | Wheat | Triticum aestivum | 20 | 2125 | [26] |
| 77 | Wild Strawberry | Fragaria vesca | 14 | 240 | [37] |
| Sr. # | Specie Name | Gene | Function | Reference |
|---|---|---|---|---|
| 1 | Arabidopsis thaliana | AtCPK1 | Cellular homeostasis, resistance fungal elicitor. | [76,78,114,115,116] |
| 2 | AtCPK3 | Salt resistance. | [117,118] | |
| 3 | AtCPK4 | Regulate ABA-regulatory transcription factors (e.g., ABF, ABF4, drought resistance). | [98] | |
| 4 | AtCPK5 | Regulate immunity responses, ROS-dependent cell-to-cell communication. | [78] | |
| 5 | AtCPK6 | Drought tolerance, ABA-dependent osmotic adjustment. | [119] | |
| 6 | AtCPK8 | Drought tolerance through interaction with protein CAT3. | [97,109] | |
| 7 | AtCPK9 | Regulate the ABA-dependent signaling pathway interacting with CPK33. | [75] | |
| 8 | AtCPK10 | Drought responsiveness, ABA-mediated stomatal movements. | [106] | |
| 9 | AtCPK11 | Phosphorylation of AtDi19, ABA signaling. | [120] | |
| 10 | AtCPK12 | Seed germination, activation of ABA regulators. | [72,121] | |
| 11 | AtCPK16 | Root-gravitropism phosphorylate AtACS7. | [122] | |
| 12 | AtCPK21 | Hyperosmotic adjustments. | [123] | |
| 13 | AtCPK23 | Salt stress, drought stress. | [124] | |
| 14 | AtCPK27 | Salinity resistance, H2O2 and ionic homeostasis. | [125] | |
| 15 | AtCPK28 | Vascular development, stem elongation, ethylene synthesis, lignin deposition. | [81,82] | |
| 16 | AtCPK32 | ABA-regulatory gene activation. | [126] | |
| 17 | AtCPK33 | Regulates flowering, biosynthesis of florigen and flowering locus T protein. | [127] | |
| 18 | Cicer areitinum (Chickpea) | CaCPK1 | Salt stress, drought stress, phytohormones, and defense signaling pathways. | [110] |
| 19 | CaCPK2 | |||
| 20 | Capsicum annuum (Peppers) | CaCPK3 | Pathogen resistance, defense functioning (i.e., regulates jasmonic and salicylic acid), ethephon. | [79] |
| 21 | Fragaria x ananassa (Garden strawberry) | FaCPK1 | low-temperature tolerance, fruit ripening. | [128] |
| 22 | Medicago sativa (Alfalfa) | MsCPK3 | Heat stress resistance, embryogenesis. | [129] |
| 23 | Oryza sativa (Rice) | OsCPK1 | Drought stress, seed germination, and GA biosynthesis. | [111] |
| 24 | OsCPK4 | Microbial-associated immunity, OsRLCK176 degradation. | [130] | |
| 25 | OsCDPK5 | Fungal attacks phosphorylate OsERG1 and OsERG3. | [131] | |
| 26 | OsCPK9 | Drought stress tolerance, ABA sensitivity spikelet fertility. | [71] | |
| 27 | OsCPK10 | Pseudomonas syringae pv resistance, SA and JA regulator. | [132] | |
| 28 | OsCPK12 | Salt tolerance, blast disease resistance, induce ROS production, leaf senescence, | [1,133] | |
| 29 | OsCDPK13 | Regulate cold, salt, dehydration responses. | [134] | |
| 30 | OsCPK17 | Cold stress interacts with sucrose synthase and plasma membrane intrinsic proteins. | [135] | |
| 31 | OsCPK21 | Salt tolerance, ABA pathway activation. | [136] | |
| 32 | OsCPK24 | Cold stress tolerance, inhibition of OsGrx10. | [74] | |
| 33 | OsCPK31 | Starch accumulation, early grain filling. | [137] | |
| 34 | Nicotiana tabacum (Tobacco) | NtCPK1 | Signaling localization for repression of shoot growth, GA biosynthesis. | [138] |
| 35 | NtCPK2 | Biotic stress immunity. | [139] | |
| 36 | NtCPK32 | Pollen tube growth interacts with CNGC18. | [83] | |
| 37 | Hevea brasiliensis (Rubber tree) | HbCDPK1 | Latex biosynthesis, rubber production. | [84] |
| 38 | Panax ginseng (Chinese ginseng) | PgCDPK1a | Regulate ginseng growth. | [85] |
| 39 | Phalaenopsis amabilis (Moth orchid) | PaCPK1 | Cold stress sensitivity, wounding, pathogen attack. | [86] |
| 40 | Triticum aestivum (Wheat) | TaCDPK1 | Regulate metabolic and developmental pathways. | [91] |
| 41 | TaCPK7 | Drought stress, salt stress, ABA signaling pathway. | [112] | |
| 42 | TaCPK12 | |||
| 43 | Zingiber officinale (Ginger) | ZoCDPK1 | Salinity and drought stress tolerance. | [113] |
| 44 | Zea mays (Maize) | ZmCPK1 | Cold stress regulates ZmERF3 expression. | [33] |
| 45 | ZmCPK4 | Upregulate ABA-regulatory components (i.e., ABI5, ABF3 and RAB18) with MAPKs. | [87] | |
| 46 | ZmCPK11 | Superoxide dismutase and ascorbate peroxidase production, ABA pathway. | [103] | |
| 47 | Vigna radiata (Mung bean) | VrCPK1 | Salt stress tolerance. | [140] |
| 48 | Vicia faba (Broad bean) | VfCPK1 | Drought stress resistance. | [108] |
| 49 | Solanum lycopersicum (Tomato) | SlCDPK2 | Flowering. | [141] |
| 50 | SlCDPK10 | Xanthomonas oryzae pv. oryzae and Pseudomonas syringae resistance. | ||
| 51 | SlCDPK18 | Xanthomonas oryzae pv. oryzae and Pseudomonas syringae resistance. | [60] | |
| 52 | Solanum tuberosum (Potato) | StCPK1 | Tuber formation. | [142] |
| 53 | StCPK4 | Fungal pathogen resistance, ROS production. | [143] | |
| 54 | StCDPK5 | Blight resistance and susceptibility, ROS defense functioning. | [100] | |
| 55 | StCDPK7 | Resistance against Phytophthora infestans. | [77] | |
| 56 | Nicotiana attenuate (Coyote tobacco) | NaCDPK4 | Wound-induced jasmonic acid (JA) accumulation, insect resistance. | [80] |
| 57 | NaCDPK5 | |||
| 58 | Camellia sinensis (Tea plant) | CsCDPK20 | High-temperature stress resistance. | [144] |
| 59 | CsCDPK26 | |||
| 60 | Hordeum vulgare (Barley) | HvCPK3 | Resistance against powdery mildew. | [145] |
| 61 | HvCPK4 | |||
| 62 | Brassica napus (Oilseed rape) | BnaCPK2 | ROS accumulation, cell death. | [2] |
| 63 | Musa acuminate (Banana) | MaCDPK7 | Heat-induced fruit ripening, chilling, stress tolerance. | [146] |
| 64 | MaCDPK2 | Sensitive to Foc-TR4 infection, biotic stress tolerance. | [147] | |
| 65 | MaCDPK4 | Sensitive to Foc-TR4 infection, biotic stress tolerance. | ||
| 66 | MaCDPK3 | Responsive for drought, cold, and salinity. | ||
| 67 | Vitis amurensis (Grapevine) | VaCPK1 | Salt stress, heat-responsiveness, stilbene bio-synthesis. | [89,148] |
| 68 | VaCPK26 | Salt stress, Stilbene bio-synthesis, through the induced expression of stilbene synthase (STS) genes. | [89,148] | |
| 69 | VaCPK20 | Drought stress, cold stress. | [107] | |
| 70 | VaCPK21 | Salt stress signaling. | [149] | |
| 71 | Pharbitis nil (Picotee) | PnCPK1 | Seed germination, seedling growth, flowering, regulation of light-dependent pathways, embryogenesis. | [90] |
| 72 | Populus euphratica (Desert poplar) | PeCPK10 | Drought and cold stress tolerance, ABA-responsive genes regulator. | [150] |
| 73 | Cucumis melo (Hami melon) | HmCDPK2 | Resistance against Penicillium infection. | [151] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atif, R.M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M.A.R.; Azeem, F.; Nawaz, M.A.; Wani, S.H.; Chung, G. Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2019, 20, 5298. https://doi.org/10.3390/ijms20215298
Atif RM, Shahid L, Waqas M, Ali B, Rashid MAR, Azeem F, Nawaz MA, Wani SH, Chung G. Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. International Journal of Molecular Sciences. 2019; 20(21):5298. https://doi.org/10.3390/ijms20215298
Chicago/Turabian StyleAtif, Rana Muhammad, Luqman Shahid, Muhammad Waqas, Babar Ali, Muhammad Abdul Rehman Rashid, Farrukh Azeem, Muhammad Amjad Nawaz, Shabir Hussain Wani, and Gyuhwa Chung. 2019. "Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants" International Journal of Molecular Sciences 20, no. 21: 5298. https://doi.org/10.3390/ijms20215298
APA StyleAtif, R. M., Shahid, L., Waqas, M., Ali, B., Rashid, M. A. R., Azeem, F., Nawaz, M. A., Wani, S. H., & Chung, G. (2019). Insights on Calcium-Dependent Protein Kinases (CPKs) Signaling for Abiotic Stress Tolerance in Plants. International Journal of Molecular Sciences, 20(21), 5298. https://doi.org/10.3390/ijms20215298