Immunomodulatory Protein from Nectria haematococca Induces Apoptosis in Lung Cancer Cells via the P53 Pathway
Abstract
:1. Introduction
2. Results
2.1. Recombinant FIP-Nha (rFIP-nha) has Stronger Antitumor Capacity Than the Widely Studied FIPs
2.2. Metabolite Profile Analysis by 1H NMR Metabolomics
2.3. Metabolic Pathway Analysis
2.4. Validation of P53 Expression
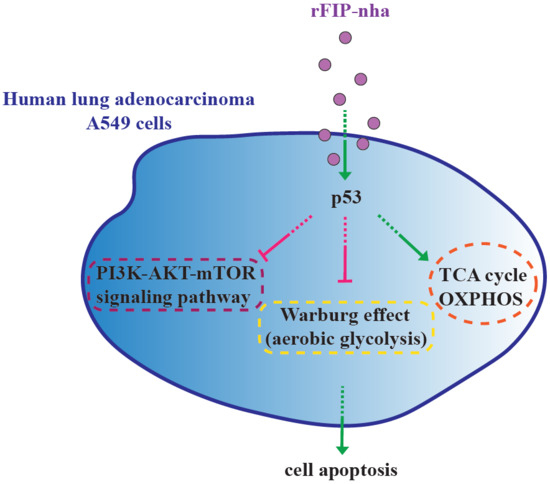
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of FIPs
4.2. Cell Culture
4.3. Tumor Cell Proliferative Inhibitory Assay
4.4. Sample Preparation
4.5. 1H NMR Measurements and Spectral Data Processing
4.6. NMR Metabolomics Data Analysis with MetaboAnalyst
4.7. Real-Time PCR Assay for P53 Expression
4.8. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.I.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Ma, Z.; Li, A.; Li, H.; Wang, B.; Zhong, J.; Min, L.; Dai, L. Metabolomic profiling of human serum in lung cancer patients using liquid chromatography/hybrid quadrupole time-of-flight mass spectrometry and gas chromatography/mass spectrometry. J. Cancer Res. Clin. Oncol. 2015, 141, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Baird, R.D.; Kaye, S.B. Drug resistance reversal—Are we getting closer? Eur. J. Cancer 2003, 39, 2450–2461. [Google Scholar] [CrossRef]
- Kino, K.; Yamashita, A.; Yamaoka, K.; Watanabe, J.; Tanaka, S.; Ko, K.; Shimizu, K.; Tsunoo, H. Isolation and characterization of a new immunomodulatory protein, ling zhi-8 (LZ-8), from Ganoderma lucidium. J. Biol. Chem. 1989, 264, 472–478. [Google Scholar] [PubMed]
- Li, J.; Cheng, C.; Yang, W.; Yang, C.; Ou, Y.; Wu, M.; Ko, J. FIP-gts Potentiate Autophagic Cell Death Against Cisplatin-resistant Urothelial Cancer Cells. Anticancer Res. 2014, 34, 2973–2983. [Google Scholar]
- Chiu, L.; Hu, M.; Yang, T.; Hsin, I.; Ko, J.; Tsai, K.; Sheu, G. Immunomodulatory Protein from Ganoderma microsporum Induces Pro-Death Autophagy through Akt-mTOR-p70S6K Pathway Inhibition in Multidrug Resistant Lung Cancer Cells. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Carrola, J.; Rocha, C.M.; Barros, A.S.; Gil, A.M.; Goodfellow, B.J.; Carreira, I.M.; Bernardo, J.; Gomes, A.; Sousa, V.; Carvalho, L. Metabolic Signatures of Lung Cancer in Biofluids: NMR-Based Metabonomics of Urine. J. Proteome Res. 2011, 10, 221–230. [Google Scholar] [CrossRef]
- Wang, X.F.; Su, K.Q.; Bao, T.W.; Cong, W.R.; Chen, Y.F.; Li, Q.Z.; Zhou, X.W. Immunomodulatory Effects Of Fungal Proteins. Curr. Top. Nutraceuticals Res. 2012, 10, 1–11. [Google Scholar]
- Paaventhan, P.; Joseph, J.S.; Seow, S.V.; Vaday, S.; Robinson, H.; Chua, K.Y.; Kolatkar, P.R. A 1.7Å Structure of Fve, a Member of the New Fungal Immunomodulatory Protein Family. J. Mol. Biol. 2003, 332, 461–470. [Google Scholar] [CrossRef]
- Huang, L.; Sun, F.; Liang, C.; He, Y.X.; Bao, R.; Liu, L.; Zhou, C.Z. Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium. Proteins 2009, 75, 524–527. [Google Scholar] [CrossRef]
- Tennant, D.A.; Duran, R.V.; Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 2010, 10, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Chen, J.; Gao, Y.; Sun, J.; Zhang, Q.; Zhang, M.; Xu, F.; Zhang, Z. Metabolomics based on liquid chromatography with mass spectrometry reveals the chemical difference in the stems and roots derived from Ephedra sinica. J. Sep. Sci. 2015, 38, 3331–3336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Wu, R.; Liang, Y.; Lin, M.; Liu, J.; Chan, C.S.; Hu, W.; Feng, Z. Tumor suppressor p53 negatively regulates glycolysis stimulated by hypoxia through its target RRAD. Oncotarget 2014, 5, 5535–5546. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, C.; Chang, Y.; Lu, X.; Zhang, J.; Zhao, Y.; Zhao, J.; Xu, G. Metabolomics study of cured tobacco using liquid chromatography with mass spectrometry: Method development and its application in investigating the chemical differences of tobacco from three growing regions. J. Sep. Sci. 2014, 37, 1067–1074. [Google Scholar] [CrossRef]
- Wang, T.; Shao, K.; Chu, Q.; Ren, Y.; Mu, Y.; Qu, L.; He, J.; Jin, C.; Xia, B. Automics: An integrated platform for NMR-based metabonomics spectral processing and data analysis. BMC Bioinform. 2009, 10, 83. [Google Scholar] [CrossRef]
- An, D.; Zhang, Q.; Wu, S.; Wei, J.; Yang, J.; Dong, F.; Yan, X.; Guo, C. Changes of metabolic profiles in urine after oral administration of quercetin in rats. Food Chem. Toxicol. 2010, 48, 1521–1527. [Google Scholar] [CrossRef]
- Griffin, J.L. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr. Opin. Chem. Biol. 2003, 7, 648–654. [Google Scholar] [CrossRef]
- Feng, J.; Zhao, J.; Hao, F.; Chen, C.; Bhakoo, K.; Tang, H. NMR-based metabonomic analyses of the effects of ultrasmall superparamagnetic particles of iron oxide (USPIO) on macrophage metabolism. J. Nanoparticle Res. 2011, 13, 2049–2062. [Google Scholar] [CrossRef]
- Li, S.; Nie, Y.; Ding, Y.; Shi, L.; Tang, X. Recombinant Expression of a Novel Fungal Immunomodulatory Protein with Human Tumor Cell Antiproliferative Activity from Nectria haematococca. Int. J. Mol. Sci. 2014, 15, 17751–17764. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Li, S.; Sun, L.; Liu, S.; Wang, F.; Wen, B.; Sun, L.; Fang, X.; Chai, Y.; Cao, H. Fungal Immunomodulatory Protein from Nectria haematococca Suppresses Growth of Human Lung Adenocarcinoma by Inhibiting the PI3K/Akt Pathway. Int. J. Mol. Sci. 2018, 19, 3429. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wang, X.; Mancuso, A.; Gao, X.; Wu, M.; Yang, X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011, 13, 310–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Hsiao, Y.; Wu, M.; Ou, C.; Lin, Y.; Lue, K.; Ko, J. Interruption of Lung Cancer Cell Migration and Proliferation by Fungal Immunomodulatory Protein FIP-fve from Flammulina velutipes. J. Agric. Food Chem. 2013, 61, 12044–12052. [Google Scholar] [CrossRef] [PubMed]
- Rankin, N.; Preiss, D.; Welsh, P.; Burgess, K.E.V.; Nelson, S.M.; Lawlor, D.A.; Sattar, N. The emergence of proton nuclear magnetic resonance metabolomics in the cardiovascular arena as viewed from a clinical perspective. Atherosclerosis 2014, 237, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Mao, Y.; Zhao, Y.; Cao, Y.; Chen, X. Role of multifaceted regulators in cancer glucose metabolism and their clinical significance. Oncotarget 2016, 7, 31572–31585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Chen, X.; Wang, L.; Chen, S. The sweet trap in tumors: Aerobic glycolysis and potential targets for therapy. Oncotarget 2016, 7, 38908–38926. [Google Scholar] [CrossRef] [PubMed]
- Hsin, I.; Ou, C.; Wu, T.; Jan, M.; Wu, M.; Chiu, L.; Lue, K.; Ko, J. GMI, an immunomodulatory protein from Ganoderma microsporum, induces autophagy in non-small cell lung cancer cells. Autophagy 2011, 7, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Z.; Sun, L.; Liu, X.; Huang, Y.; Wang, F.; Xin, F. Characterization of a new fungal immunomodulatory protein, FIP-dsq2 from Dichomitus squalens. J. Biotechnol. 2017, 246, 45–51. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Z.; Xu, W.; Xie, Y.; Zhao, L.; Tang, X.; Wang, F.; Xin, F. FIP-sch2, a new fungal immunomodulatory protein from Stachybotrys chlorohalonata, suppresses proliferation and migration in lung cancer cells. Appl. Microbiol. Biotechnol. 2017, 101, 3227–3235. [Google Scholar] [CrossRef]
- Li, S.Y.; Shi, L.J.; Ding, Y.; Nie, Y.; Tang, X.M. Identification and functional characterization of a novel fungal immunomodulatory protein from Postia placenta. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2015, 78, 64–70. [Google Scholar] [CrossRef]
- Cong, W.; Xu, H.; Liu, Y.; Li, Q.; Li, W.; Zhou, X. Production and functional characterization of a novel fungal immunomodulatory protein FIP-SN15 shuffled from two genes of Ganoderma species. Appl. Microbiol. Biotechnol. 2014, 98, 5967–5975. [Google Scholar] [CrossRef]
- Liao, C.; Hsiao, Y.; Lin, C.; Yeh, C.; Wang, J.C.; Ni, C.; Hsu, C.; Ko, J. Induction of premature senescence in human lung cancer by fungal immunomodulatory protein from Ganoderma tsugae. Food Chem. Toxicol. 2008, 46, 1851–1859. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Hsiao, Y.; Hsu, C.; Lin, M.; Wang, J.C.; Huang, Y.; Ko, J. Transcriptionally Mediated Inhibition of Telomerase of Fungal Immunomodulatory Protein From Ganoderma tsugae in A549 Human Lung Adenocarcinoma Cell Line. Mol. Carcinog. 2006, 45, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Hsiao, Y.; Sheu, G.; Chang, J.T.; Wang, P.; Wu, M.; Shieh, G.; Hsu, C.; Ko, J. Nuclear translocation of telomerase reverse transcriptase and calcium signaling in repression of telomerase activity in human lung cancer cells by fungal immunomodulatory protein from Ganoderma tsugae. Biochem. Pharmacol. 2007, 74, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gu, W. To be, or not to be: Functional dilemma of p53 metabolic regulation. Curr. Opin. Oncol. 2014, 26, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Madan, E.; Gogna, R.; Bhatt, M.; Pati, U.; Kuppusamy, P.; Mahdi, A.A. Regulation of glucose metabolism by p53: Emerging new roles for the tumor suppressor. Oncotarget. 2011, 2, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qin, Z.; Wang, J.L. The role of p53 in cell metabolism. Acta. Pharmacol. Sin. 2010, 31, 1208–1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Liu, J.; Feng, Z. The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci. 2013, 3, 9. [Google Scholar] [CrossRef]
- Schwartzenbergbaryoseph, F.; Armoni, M.; Karnieli, E. The Tumor Suppressor p53 Down-Regulates Glucose Transporters GLUT1 and GLUT4 Gene Expression. Cancer Res. 2004, 64, 2627–2633. [Google Scholar] [CrossRef]
- Kawauchi, K.; Araki, K.; Tobiume, K.; Tanaka, N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat. Cell Biol. 2008, 10, 611–618. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Jung, H.; Lim, C. Reactive Oxygen Species-Dependent Down-Regulation of Tumor Suppressor Genes PTEN, USP28, DRAM, TIGAR, and CYLD Under Oxidative Stress. Biochem. Genet. 2013, 51, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, H.; Lleonart, M.E.; Gil, J.; Wang, J.; Degan, P.; Peters, G.; Martinez, D.; Carnero, A.; Beach, D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005, 65, 177–185. [Google Scholar] [PubMed]
- Desideri, E.; Vegliante, R.; Ciriolo, M.R. Mitochondrial dysfunctions in cancer: Genetic defects and oncogenic signaling impinging on TCA cycle activity. Cancer Lett. 2015, 356, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Lin, M.; Zhu, W.; Liang, Y.; Hong, X.; Zhao, Y.; Young, K.H.; Hu, W.; Feng, Z. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 2014, 5, 2635–2647. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Hu, W.; De Stanchina, E.; Teresky, A.K.; Jin, S.; Lowe, S.W.; Levine, A.J. The Regulation of AMPK β1, TSC2, and PTEN Expression by p53: Stress, Cell and Tissue Specificity, and the Role of These Gene Products in Modulating the IGF-1-AKT-mTOR Pathways. Cancer Res. 2007, 67, 3043–3053. [Google Scholar] [CrossRef]
- Budanov, A.V.; Karin, M. p53 Target Genes Sestrin1 and Sestrin2 Connect Genotoxic Stress and mTOR Signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef] [Green Version]






| Metabolite | r 1 | VIP 2 |
|---|---|---|
| 1-Methylnicotinamide: 8.69(m), 8.76(m), 8.82(d), 9.20(s) | 0.609 | 1.294 |
| 7-Methylguanosine: 9.08(s) | 0.501 | 1.080 |
| Acetate: 1.92(s) | −0.572 | 1.330 |
| Butyrate: 0.90(t), 2.14(t) | −0.633 | 1.411 |
| Citrate: 2.54(d), 2.70(d) | −0.856 | 1.802 |
| Creatine: 3.04(s), 3.93(s) | −0.885 | 1.856 |
| Creatinine: 3.05(s), 4.06(s) | −0.682 | 1.496 |
| Glutamine: 2.09(m), 2.43(m), 3.78(t) | −0.833 | 1.732 |
| Glycerol: 3.58(m), 3.64(m), 3.81(m) | −0.682 | 1.438 |
| Histidine: 6.98(s), 7.91(s) | 0.675 | 1.406 |
| Lactate: 1.33(d), 4.12(q) | −0.672 | 1.399 |
| Mannitol: 3.68(dd), 3.76(m), 3.81(d), 3.88(dd) | 0.913 | 1.899 |
| p-Aminobenzoate: 6.83(d), 7.71(d) | 0.676 | 1.493 |
| Phenylalanine: 7.33(d), 7.37(t), 7.43(m) | −0.632 | 1.372 |
| Pyruvate: 2.38(s) | −0.658 | 1.440 |
| Taurine: 3.27(t), 3.43(t) | −0.516 | 1.224 |
| Trimethylamine N-oxide: 3.27(s) | −0.867 | 1.804 |
| α-Glucose: 3.42(t), 3.54(dd), 3.71(t), 3.84(m), 5.24(d) | −0.875 | 1.825 |
| β-Glucose: 3.25(dd), 3.41(t), 3.46(m), 3.49(t), 3.90(dd), 4.65(d) | −0.917 | 1.907 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-J.; Wang, Y.; Hou, L.; Xin, F.; Fan, B.; Lu, C.; Zhang, L.; Wang, F.; Li, S. Immunomodulatory Protein from Nectria haematococca Induces Apoptosis in Lung Cancer Cells via the P53 Pathway. Int. J. Mol. Sci. 2019, 20, 5348. https://doi.org/10.3390/ijms20215348
Wang J-J, Wang Y, Hou L, Xin F, Fan B, Lu C, Zhang L, Wang F, Li S. Immunomodulatory Protein from Nectria haematococca Induces Apoptosis in Lung Cancer Cells via the P53 Pathway. International Journal of Molecular Sciences. 2019; 20(21):5348. https://doi.org/10.3390/ijms20215348
Chicago/Turabian StyleWang, Jing-Jing, Yan Wang, Lizhen Hou, Fengjiao Xin, Bei Fan, Cong Lu, Lijing Zhang, Fengzhong Wang, and Shuying Li. 2019. "Immunomodulatory Protein from Nectria haematococca Induces Apoptosis in Lung Cancer Cells via the P53 Pathway" International Journal of Molecular Sciences 20, no. 21: 5348. https://doi.org/10.3390/ijms20215348
APA StyleWang, J. -J., Wang, Y., Hou, L., Xin, F., Fan, B., Lu, C., Zhang, L., Wang, F., & Li, S. (2019). Immunomodulatory Protein from Nectria haematococca Induces Apoptosis in Lung Cancer Cells via the P53 Pathway. International Journal of Molecular Sciences, 20(21), 5348. https://doi.org/10.3390/ijms20215348