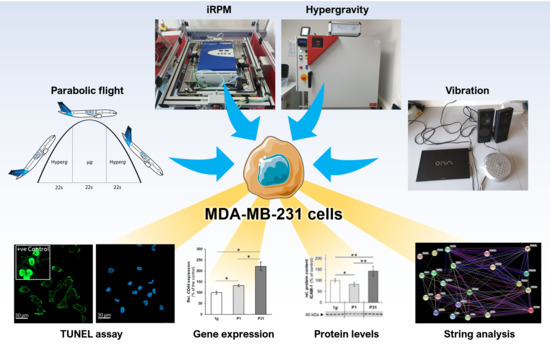
Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Viability Staining
2.2. Nuclear Factor ”Kappa-Light-Chain-Enhancer” of Activated B-Cells (NF-κB) Signaling Factors in Triple-Negative Breast Cancer Cells during Altered Gravity Conditions
2.3. Expression of Factors Belonging to the Biological Process of Apoptosis
2.4. Regulation of Cell Adhesion Molecules in Triple-Negative Breast Cancer (TNBC) Cells
2.5. Factors of the Mitogen-Activated Protein Kinase (MAPK) Signaling Pathway Known to be Involved in Cancer Progression and Metastasis
2.6. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) Analysis
3. Discussion
3.1. Influence of Altered Gravity Conditions and Vibration on Cell Survival
3.2. Impact of Real Microgravity on NF-κB Signalling in TNBC
3.3. Parabolic Flight Maneuvers Changed Cell Adhesion Factors
3.4. Interaction Network of Selected Genes Evaluated by STRING Analysis
3.5. MAPK Signaling Factors Involved in Cancer Progression and Metastasis
4. Materials and Methods
4.1. Cell Culture
4.2. 29th. Parabolic Flight Campaign
4.3. Vibration Experiments
4.4. Hyper-g Experiments
4.5. Incubator Random Positioning Machine (iRPM)
4.6. RNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
4.7. Western Blotting
4.8. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Staining
4.9. STRING Analysis
4.10. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| ANXA1 | Annexin A1 |
| ANXA2 | Annexin A2 |
| AR | Androgen receptor |
| ATCC | American Type Culture Collection |
| BAX | BCL2 Associated X, Apoptosis Regulator |
| BCL2 | B-cell lymphoma 2 |
| CASP3 | Caspase3 |
| CD44 | Cell surface glycoprotein CD44 |
| Cx+ | Connexin positive |
| CXCL12 | C-X-C Motif Chemokine Ligand 12 |
| CYC1 | Cytochrome c1 |
| DLR | Deutsches Zentrum für Luft- und Raumfahrt |
| ECM | Extracellular matrix |
| ER | Estrogen receptor |
| ERK1,2 | Extracellular Signal-Regulated Kinase 1, 2 |
| FAK1 | Focal adhesion kinase 1 |
| FCS | Fetal calf serum |
| FTC-133 | Human follicular thyroid carcinoma cell line |
| GLOBOCAN | Global Cancer Observatory |
| HARV | High-aspect ratio vessel |
| HER2 | Human epidermal growth receptor 2 |
| hyper-g | Hypergravity |
| ICAM1 | Intercellular adhesion molecule 1 |
| IKBKG | NF-kappa-B essential modulator (NEMO) |
| IKK | IκB kinase |
| iRPM | Incubator random positioning machine |
| ISS | International Space Station |
| IκB | Inhibitor of κB |
| IκBα | Inhibitor of κB alpha |
| IκBβ | Inhibitor of κB beta |
| IκBγ | Inhibitor of κB gamma |
| IκBε | Inhibitor of κB epsilon |
| JNK1 | Mitogen-activated protein kinase 8 |
| KI67 | Antigen KI67 |
| µg | Microgravity |
| MAPK | Mitogen-Activated Protein Kinase |
| MCF-7 | Michigan Cancer Foundation-7 cell line |
| MCS | Multicellular spheroids |
| MDA-MB-231 | M.D. Anderson-Metastasis Breast cancer cell line |
| ML1 | Human thyroid carcinoma cell line |
| MME | Membrane Metalloendopeptidase |
| MMP11 | Matrix Metallopeptidase 11 |
| MMP2 | Matrix Metallopeptidase 2 |
| MuSIC | Multi-sample incubator centrifuge |
| NFKB1 | NF-kappa-B transcription complex P105/P50 |
| NFKB2 | NF-kappa-B transcription complex P100/P52 |
| NFKB3/NFKB P65 | NF-kappa-B transcription complex P65 |
| NFKBIA | NF-κB-inhibitor-alpha |
| NFKBIB | NF-κB-inhibitor-beta |
| NFKBIE | NF-κB-inhibitor-epsilon |
| NFκB | Nuclear factor-kappaB |
| P | Parabola |
| PF | Parabolic flight |
| PFA | Paraformaldehyde |
| PR | Progesterone receptor |
| PRKCA | Protein kinase C alpha |
| PTK2 | Protein tyrosine kinase 2 (known as FAK1) |
| qPCR | Quantitative polymerase chain reaction |
| RAF1 | RAF proto-oncogene serine/threonine-protein kinase |
| r-µg | Real microgravity |
| RELA | V-Rel Avian Reticuloendotheliosis Viral Oncogene Homolog A |
| RPM | Random positioning machine |
| s-µg | Simulated microgravity |
| SPP1/OPN | Osteopontin |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| TBP | TATA-box binding protein |
| TNBC | Triple-negative breast cancer |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
| VCAM1 | Vascular cell adhesion protein 1 |
| VIB | Vibration |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Van’t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Soliman, N.A.; Yussif, S.M. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol. Med. 2016, 13, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Farmer, P.; Bonnefoi, H.; Becette, V.; Tubiana-Hulin, M.; Fumoleau, P.; Larsimont, D.; Macgrogan, G.; Bergh, J.; Cameron, D.; Goldstein, D.; et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005, 24, 4660–4671. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, T.B.; Knutsson, M.L.; Wehland, M.; Laursen, B.E.; Grimm, D.; Warnke, E.; Magnusson, N.E. Anti-vascular endothelial growth factor therapy in breast cancer. Int. J. Mol. Sci. 2014, 15, 23024–23041. [Google Scholar] [CrossRef]
- Cailleau, R.; Young, R.; Olive, M.; Reeves, W.J., Jr. Breast tumor cell lines from pleural effusions. J. Natl. Cancer Inst. 1974, 53, 661–674. [Google Scholar] [CrossRef]
- Becker, J.L.; Souza, G.R. Using space-based investigations to inform cancer research on Earth. Nat. Rev. Cancer 2013, 13, 315–327. [Google Scholar] [CrossRef]
- Obermaier, C.; Jankowski, V.; Schmutzler, C.; Bauer, J.; Wildgruber, R.; Infanger, M.; Kohrle, J.; Krause, E.; Weber, G.; Grimm, D. Free-flow isoelectric focusing of proteins remaining in cell fragments following sonication of thyroid carcinoma cells. Electrophoresis 2005, 26, 2109–2116. [Google Scholar] [CrossRef]
- Pietsch, J.; Kussian, R.; Sickmann, A.; Bauer, J.; Weber, G.; Nissum, M.; Westphal, K.; Egli, M.; Grosse, J.; Schonberger, J.; et al. Application of free-flow IEF to identify protein candidates changing under microgravity conditions. Proteomics 2010, 10, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Kopp, S.; Schlagberger, E.M.; Grosse, J.; Sahana, J.; Riwaldt, S.; Wehland, M.; Luetzenberg, R.; Infanger, M.; Grimm, D. Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine. Int. J. Mol. Sci. 2017, 18, 546. [Google Scholar] [CrossRef] [PubMed]
- Sahana, J.; Nassef, M.Z.; Wehland, M.; Kopp, S.; Kruger, M.; Corydon, T.J.; Infanger, M.; Bauer, J.; Grimm, D. Decreased E-Cadherin in MCF7 Human Breast Cancer Cells Forming Multicellular Spheroids Exposed to Simulated Microgravity. Proteomics 2018, 18, e1800015. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Egli, M.; Kruger, M.; Riwaldt, S.; Corydon, T.J.; Kopp, S.; Wehland, M.; Wise, P.; Infanger, M.; Mann, V.; et al. Tissue Engineering Under Microgravity Conditions-Use of Stem Cells and Specialized Cells. Stem Cells Dev. 2018, 27, 787–804. [Google Scholar] [CrossRef]
- Masiello, M.G.; Cucina, A.; Proietti, S.; Palombo, A.; Coluccia, P.; D’Anselmi, F.; Dinicola, S.; Pasqualato, A.; Morini, V.; Bizzarri, M. Phenotypic switch induced by simulated microgravity on MDA-MB-231 breast cancer cells. BioMed Res. Int. 2014, 2014, 652434. [Google Scholar] [CrossRef]
- Kruger, M.; Melnik, D.; Kopp, S.; Buken, C.; Sahana, J.; Bauer, J.; Wehland, M.; Hemmersbach, R.; Corydon, T.J.; Infanger, M.; et al. Fighting Thyroid Cancer with Microgravity Research. Int. J. Mol. Sci. 2019, 20, 2553. [Google Scholar] [CrossRef]
- Wang, W.; Nag, S.A.; Zhang, R. Targeting the NFkappaB signaling pathways for breast cancer prevention and therapy. Curr. Med. Chem. 2015, 22, 264–289. [Google Scholar] [CrossRef]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef]
- Grosse, J.; Wehland, M.; Pietsch, J.; Schulz, H.; Saar, K.; Hubner, N.; Eilles, C.; Bauer, J.; Abou-El-Ardat, K.; Baatout, S.; et al. Gravity-sensitive signaling drives 3-dimensional formation of multicellular thyroid cancer spheroids. FASEB J. 2012, 26, 5124–5140. [Google Scholar] [CrossRef]
- Kopp, S.; Sahana, J.; Islam, T.; Petersen, A.G.; Bauer, J.; Corydon, T.J.; Schulz, H.; Saar, K.; Huebner, N.; Slumstrup, L.; et al. The role of NFkappaB in spheroid formation of human breast cancer cells cultured on the Random Positioning Machine. Sci. Rep. 2018, 8, 921. [Google Scholar] [CrossRef]
- Murakami, N.; Kuhnel, A.; Schmid, T.E.; Ilicic, K.; Stangl, S.; Braun, I.S.; Gehrmann, M.; Molls, M.; Itami, J.; Multhoff, G. Role of membrane Hsp70 in radiation sensitivity of tumor cells. Radiat. Oncol. 2015, 10, 149. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef]
- Huang, T.; Kang, W.; Zhang, B.; Wu, F.; Dong, Y.; Tong, J.H.; Yang, W.; Zhou, Y.; Zhang, L.; Cheng, A.S.; et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-kappaB signaling in gastric carcinogenesis. Mol Cancer 2016, 15, 9. [Google Scholar] [CrossRef]
- Lindner, V. The NF-kappaB and IkappaB system in injured arteries. Pathobiology 1998, 66, 311–320. [Google Scholar] [CrossRef]
- Bist, P.; Leow, S.C.; Phua, Q.H.; Shu, S.; Zhuang, Q.; Loh, W.T.; Nguyen, T.H.; Zhou, J.B.; Hooi, S.C.; Lim, L.H. Annexin-1 interacts with NEMO and RIP1 to constitutively activate IKK complex and NF-kappaB: Implication in breast cancer metastasis. Oncogene 2011, 30, 3174–3185. [Google Scholar] [CrossRef]
- Anwar, K.N.; Fazal, F.; Malik, A.B.; Rahman, A. RhoA/Rho-associated kinase pathway selectively regulates thrombin-induced intercellular adhesion molecule-1 expression in endothelial cells via activation of I kappa B kinase beta and phosphorylation of RelA/p65. J. Immunol. 2004, 173, 6965–6972. [Google Scholar] [CrossRef]
- Ortiz-Martinez, F.; Sanmartin, E.; Pomares-Navarro, E.; Perez-Balaguer, A.; Andres, L.; Sanchez-Paya, J.; Aranda, F.I.; Lerma, E.; Peiro, G. Osteopontin Regulates VEGFA and ICAM-1 mRNA Expression in Breast Carcinoma. Am. J. Clin. Pathol. 2015, 143, 812–822. [Google Scholar] [CrossRef]
- Ue, T.; Yokozaki, H.; Kitadai, Y.; Yamamoto, S.; Yasui, W.; Ishikawa, T.; Tahara, E. Co-expression of osteopontin and CD44v9 in gastric cancer. Int. J. Cancer 1998, 79, 127–132. [Google Scholar] [CrossRef]
- Chintagari, N.R.; Jin, N.; Wang, P.; Narasaraju, T.A.; Chen, J.; Liu, L. Effect of cholesterol depletion on exocytosis of alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 2006, 34, 677–687. [Google Scholar] [CrossRef]
- Tiys, E.S.; Ivanisenko, T.V.; Demenkov, P.S.; Ivanisenko, V.A. FunGeneNet: A web tool to estimate enrichment of functional interactions in experimental gene sets. BMC Genom. 2018, 19, 76. [Google Scholar] [CrossRef]
- Zaczynska, E.; Kochanowska, I.; Kruzel, M.; Zimecki, M. Lactoferrin Prevents Susceptibility of WEHI 231 Cells to Anti-Ig-Induced Cell Death Promoting Cell Differentiation. Folia Biol. (Praha) 2018, 64, 16–22. [Google Scholar]
- Reza, A.; Choi, Y.J.; Yasuda, H.; Kim, J.H. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Rep. 2016, 6, 38498. [Google Scholar] [CrossRef]
- Cailleau, R.; Olive, M.; Cruciger, Q.V. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef]
- Brinkley, B.R.; Beall, P.T.; Wible, L.J.; Mace, M.L.; Turner, D.S.; Cailleau, R.M. Variations in cell form and cytoskeleton in human breast carcinoma cells in vitro. Cancer Res. 1980, 40, 3118–3129. [Google Scholar]
- White, R.J.; Averner, M. Humans in space. Nature 2001, 409, 1115–1118. [Google Scholar] [CrossRef]
- Hader, D.P.; Braun, M.; Grimm, D.; Hemmersbach, R. Gravireceptors in eukaryotes-a comparison of case studies on the cellular level. NPJ Microgravity 2017, 3, 13. [Google Scholar] [CrossRef] [Green Version]
- Grimm, D.; Bauer, J.; Kossmehl, P.; Shakibaei, M.; Schoberger, J.; Pickenhahn, H.; Schulze-Tanzil, G.; Vetter, R.; Eilles, C.; Paul, M.; et al. Simulated microgravity alters differentiation and increases apoptosis in human follicular thyroid carcinoma cells. FASEB J. 2002, 16, 604–606. [Google Scholar] [CrossRef]
- Qian, A.; Zhang, W.; Xie, L.; Weng, Y.; Yang, P.; Wang, Z.; Hu, L.; Xu, H.; Tian, Z.; Shang, P. Simulated weightlessness alters biological characteristics of human breast cancer cell line MCF-7. Acta Astronaut. 2008, 63, 947–958. [Google Scholar] [CrossRef]
- Kopp, S.; Warnke, E.; Wehland, M.; Aleshcheva, G.; Magnusson, N.E.; Hemmersbach, R.; Corydon, T.J.; Bauer, J.; Infanger, M.; Grimm, D. Mechanisms of three-dimensional growth of thyroid cells during long-term simulated microgravity. Sci. Rep. 2015, 5, 16691. [Google Scholar] [CrossRef] [Green Version]
- Kopp, S.; Slumstrup, L.; Corydon, T.J.; Sahana, J.; Aleshcheva, G.; Islam, T.; Magnusson, N.E.; Wehland, M.; Bauer, J.; Infanger, M.; et al. Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci. Rep. 2016, 6, 26887. [Google Scholar] [CrossRef]
- Schoenberger, J.; Bauer, J.; Moosbauer, J.; Eilles, C.; Grimm, D. Innovative strategies in in vivo apoptosis imaging. Curr. Med. Chem. 2008, 15, 187–194. [Google Scholar] [CrossRef]
- Grosse, J.; Wehland, M.; Pietsch, J.; Ma, X.; Ulbrich, C.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; et al. Short-term weightlessness produced by parabolic flight maneuvers altered gene expression patterns in human endothelial cells. FASEB J. 2012, 26, 639–655. [Google Scholar] [CrossRef]
- Wehland, M.; Ma, X.; Braun, M.; Hauslage, J.; Hemmersbach, R.; Bauer, J.; Grosse, J.; Infanger, M.; Grimm, D. The impact of altered gravity and vibration on endothelial cells during a parabolic flight. Cell Physiol. Biochem. 2013, 31, 432–451. [Google Scholar] [CrossRef]
- Sarkar, D.K.; Jana, D.; Patil, P.S.; Chaudhari, K.S.; Chattopadhyay, B.K.; Chikkala, B.R.; Mandal, S.; Chowdhary, P. Role of NF-kappaB as a Prognostic Marker in Breast Cancer: A Pilot Study in Indian Patients. Indian J. Surg. Oncol. 2013, 4, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Khongthong, P.; Roseweir, A.K.; Edwards, J. The NF-KB pathway and endocrine therapy resistance in breast cancer. Endocr. Relat. Cancer 2019, 26, 369–380. [Google Scholar] [CrossRef]
- Ulbrich, C.; Westphal, K.; Baatout, S.; Wehland, M.; Bauer, J.; Flick, B.; Infanger, M.; Kreutz, R.; Vadrucci, S.; Egli, M.; et al. Effects of basic fibroblast growth factor on endothelial cells under conditions of simulated microgravity. J. Cell Biochem. 2008, 104, 1324–1341. [Google Scholar] [CrossRef]
- Ghobrial, I.M.; Witzig, T.E.; Adjei, A.A. Targeting apoptosis pathways in cancer therapy. CA Cancer J. Clin. 2005, 55, 178–194. [Google Scholar] [CrossRef]
- Chang, T.T.; Walther, I.; Li, C.F.; Boonyaratanakornkit, J.; Galleri, G.; Meloni, M.A.; Pippia, P.; Cogoli, A.; Hughes-Fulford, M. The Rel/NF-kappaB pathway and transcription of immediate early genes in T cell activation are inhibited by microgravity. J. Leukoc. Biol. 2012, 92, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Boonyaratanakornkit, J.B.; Cogoli, A.; Li, C.F.; Schopper, T.; Pippia, P.; Galleri, G.; Meloni, M.A.; Hughes-Fulford, M. Key gravity-sensitive signaling pathways drive T cell activation. FASEB J. 2005, 19, 2020–2022. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [Green Version]
- Wuest, S.L.; Stern, P.; Casartelli, E.; Egli, M. Fluid Dynamics Appearing during Simulated Microgravity Using Random Positioning Machines. PLoS ONE 2017, 12, e0170826. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Bauer, J.; Braun, M.; Schwarzwalder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Buken, C.; Sahana, J.; Corydon, T.J.; Melnik, D.; Bauer, J.; Wehland, M.; Kruger, M.; Balk, S.; Abuagela, N.; Infanger, M.; et al. Morphological and Molecular Changes in Juvenile Normal Human Fibroblasts Exposed to Simulated Microgravity. Sci. Rep. 2019, 9, 11882. [Google Scholar] [CrossRef] [PubMed]
- Nassef, M.Z.; Kopp, S.; Wehland, M.; Melnik, D.; Sahana, J.; Kruger, M.; Corydon, T.J.; Oltmann, H.; Schmitz, B.; Schutte, A.; et al. Real Microgravity Influences the Cytoskeleton and Focal Adhesions in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3456. [Google Scholar] [CrossRef] [Green Version]
- Dittrich, A.; Grimm, D.; Sahana, J.; Bauer, J.; Kruger, M.; Infanger, M.; Magnusson, N.E. Key Proteins Involved in Spheroid Formation and Angiogenesis in Endothelial Cells After Long-Term Exposure to Simulated Microgravity. Cell Physiol. Biochem. 2018, 45, 429–445. [Google Scholar] [CrossRef]
- Eibl, R.H.; Benoit, M. Molecular resolution of cell adhesion forces. IEE Proc. Nanobiotechnol. 2004, 151, 128–132. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, D.; Shan, S.; Wu, J.; Yang, H.; Yu, Y. Angiogenic effect of intercellular adhesion molecule-1. J. Huazhong Univ. Sci. Technol. Med. Sci. 2007, 27, 9–12. [Google Scholar] [CrossRef]
- Tauber, S.; Lauber, B.A.; Paulsen, K.; Layer, L.E.; Lehmann, M.; Hauschild, S.; Shepherd, N.R.; Polzer, J.; Segerer, J.; Thiel, C.S.; et al. Cytoskeletal stability and metabolic alterations in primary human macrophages in long-term microgravity. PLoS ONE 2017, 12, e0175599. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, K.; Tauber, S.; Dumrese, C.; Bradacs, G.; Simmet, D.M.; Golz, N.; Hauschild, S.; Raig, C.; Engeli, S.; Gutewort, A.; et al. Regulation of ICAM-1 in cells of the monocyte/macrophage system in microgravity. BioMed Res. Int. 2015, 2015, 538786. [Google Scholar] [CrossRef]
- Ulbrich, C.; Pietsch, J.; Grosse, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; Braun, M.; et al. Differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: Relationship between the extracellular matrix and the cytoskeleton. Cell Physiol. Biochem. 2011, 28, 185–198. [Google Scholar] [CrossRef]
- Aleshcheva, G.; Wehland, M.; Sahana, J.; Bauer, J.; Corydon, T.J.; Hemmersbach, R.; Frett, T.; Egli, M.; Infanger, M.; Grosse, J.; et al. Moderate alterations of the cytoskeleton in human chondrocytes after short-term microgravity produced by parabolic flight maneuvers could be prevented by up-regulation of BMP-2 and SOX-9. FASEB J. 2015, 29, 2303–2314. [Google Scholar] [CrossRef]
- Ingram, M.; Techy, G.B.; Saroufeem, R.; Yazan, O.; Narayan, K.S.; Goodwin, T.J.; Spaulding, G.F. Three-dimensional growth patterns of various human tumor cell lines in simulated microgravity of a NASA bioreactor. In Vitro Cell Dev. Biol. Anim. 1997, 33, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Kumei, Y.; Morita, S.; Katano, H.; Akiyama, H.; Hirano, M.; Oyha, K.; Shimokawa, H. Microgravity signal ensnarls cell adhesion, cytoskeleton, and matrix proteins of rat osteoblasts: Osteopontin, CD44, osteonectin, and alpha-tubulin. Ann. N. Y. Acad. Sci. 2006, 1090, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.F. The cancer biomarker osteopontin: Combination with other markers. Cancer Genom. Proteom. 2011, 8, 263–288. [Google Scholar]
- Irby, R.B.; McCarthy, S.M.; Yeatman, T.J. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clin. Exp. Metastasis 2004, 21, 515–523. [Google Scholar] [CrossRef]
- Wei, R.; Wong, J.P.C.; Kwok, H.F. Osteopontin—A promising biomarker for cancer therapy. J. Cancer 2017, 8, 2173–2183. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-kappaB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef] [Green Version]
- Weber, G.F.; Ashkar, S.; Cantor, H. Interaction between CD44 and osteopontin as a potential basis for metastasis formation. Proc. Assoc. Am. Physicians 1997, 109, 1–9. [Google Scholar]
- Ahmed, M.; Kundu, G.C. Osteopontin selectively regulates p70S6K/mTOR phosphorylation leading to NF-kappaB dependent AP-1-mediated ICAM-1 expression in breast cancer cells. Mol. Cancer 2010, 9, 101. [Google Scholar] [CrossRef] [Green Version]
- O’Hanlon, D.M.; Fitzsimons, H.; Lynch, J.; Tormey, S.; Malone, C.; Given, H.F. Soluble adhesion molecules (E-selectin, ICAM-1 and VCAM-1) in breast carcinoma. Eur. J. Cancer 2002, 38, 2252–2257. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, K.; Lee, E.; Ahn, T.; Jung, H.H.; Lim, S.H.; Hong, M.; Do, I.G.; Cho, E.Y.; Kim, D.H.; et al. Gene Expression Profiling of Breast Cancer Brain Metastasis. Sci. Rep. 2016, 6, 28623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, L.D.; Vishwanatha, J.K. Prognostic impact of AnxA1 and AnxA2 gene expression in triple-negative breast cancer. Oncotarget 2018, 9, 2697–2704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.; Kim, J.S.; Kim, W.K.; Oh, K.J.; Kim, J.M.; Lee, H.J.; Han, B.S.; Kim, D.S.; Seo, Y.S.; Lee, S.C.; et al. Intracellular annexin A2 regulates NF-kappaB signaling by binding to the p50 subunit: Implications for gemcitabine resistance in pancreatic cancer. Cell Death Dis. 2015, 6, e1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, K.M.; Dong, S.; Fan, M.; Li, J.J. Nuclear factor-kappaB p65 inhibits mitogen-activated protein kinase signaling pathway in radioresistant breast cancer cells. Mol. Cancer Res. 2006, 4, 945–955. [Google Scholar] [CrossRef] [Green Version]
- Cook, S.J.; Stuart, K.; Gilley, R.; Sale, M.J. Control of cell death and mitochondrial fission by ERK1/2 MAP kinase signalling. FEBS J. 2017, 284, 4177–4195. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.N.D.; Perez White, B.E.; Zhao, H.; Mortazavi, F.; Tonetti, D.A. Protein kinase C alpha enhances migration of breast cancer cells through FOXC2-mediated repression of p120-catenin. BMC Cancer 2017, 17, 832. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.; Shi, Y.; Yang, D.; Xu, M.; Lai, Y.; Liu, Y. Identification of key candidate genes involved in melanoma metastasis. Mol. Med. Rep. 2019, 20, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Kolch, W.; Heidecker, G.; Kochs, G.; Hummel, R.; Vahidi, H.; Mischak, H.; Finkenzeller, G.; Marme, D.; Rapp, U.R. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature 1993, 364, 249–252. [Google Scholar] [CrossRef]
- Olea-Flores, M.; Zuniga-Eulogio, M.D.; Mendoza-Catalan, M.A.; Rodriguez-Ruiz, H.A.; Castaneda-Saucedo, E.; Ortuno-Pineda, C.; Padilla-Benavides, T.; Navarro-Tito, N. Extracellular-Signal Regulated Kinase: A Central Molecule Driving Epithelial-Mesenchymal Transition in Cancer. Int. J. Mol. Sci. 2019, 20, 2885. [Google Scholar] [CrossRef] [Green Version]
- Eblen, S.T. Extracellular-Regulated Kinases: Signaling From Ras to ERK Substrates to Control Biological Outcomes. Adv. Cancer Res. 2018, 138, 99–142. [Google Scholar] [CrossRef]
- Crapoulet, N.; O’Brien, P.; Ouellette, R.J.; Robichaud, G.A. Coordinated expression of Pax-5 and FAK1 in metastasis. Anticancer Agents Med. Chem. 2011, 11, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.T.; Cortesio, C.L.; Huttenlocher, A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J. Cell Biol. 2009, 185, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.S.; Crowe, D.L. Jun amino-terminal kinase 1 activation promotes cell survival in ErbB2-positive breast cancer. Anticancer Res. 2010, 30, 3407–3412. [Google Scholar] [PubMed]
- Zhang, J.Y.; Selim, M.A. The role of the c-Jun N-terminal Kinase signaling pathway in skin cancer. Am. J. Cancer Res. 2012, 2, 691–698. [Google Scholar]
- Ashenden, M.; van Weverwijk, A.; Murugaesu, N.; Fearns, A.; Campbell, J.; Gao, Q.; Iravani, M.; Isacke, C.M. An In Vivo Functional Screen Identifies JNK Signaling As a Modulator of Chemotherapeutic Response in Breast Cancer. Mol. Cancer Ther. 2017, 16, 1967–1978. [Google Scholar] [CrossRef] [Green Version]
- Lutzenberg, R.; Solano, K.; Buken, C.; Sahana, J.; Riwaldt, S.; Kopp, S.; Kruger, M.; Schulz, H.; Saar, K.; Huebner, N.; et al. Pathway Analysis Hints Towards Beneficial Effects of Long-Term Vibration on Human Chondrocytes. Cell Physiol. Biochem. 2018, 47, 1729–1741. [Google Scholar] [CrossRef]
- Corydon, T.J.; Kopp, S.; Wehland, M.; Braun, M.; Schutte, A.; Mayer, T.; Hulsing, T.; Oltmann, H.; Schmitz, B.; Hemmersbach, R.; et al. Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci. Rep. 2016, 6, 20043. [Google Scholar] [CrossRef] [Green Version]
- Benavides Damm, T.; Walther, I.; Wuest, S.L.; Sekler, J.; Egli, M. Cell cultivation under different gravitational loads using a novel random positioning incubator. Biotechnol. Bioeng. 2014, 111, 1180–1190. [Google Scholar] [CrossRef] [Green Version]
- Lutzenberg, R.; Wehland, M.; Solano, K.; Nassef, M.Z.; Buken, C.; Melnik, D.; Bauer, J.; Kopp, S.; Kruger, M.; Riwaldt, S.; et al. Beneficial Effects of Low Frequency Vibration on Human Chondrocytes in Vitro. Cell Physiol. Biochem. 2019, 53, 623–637. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [Green Version]
- Pietsch, J.; Riwaldt, S.; Bauer, J.; Sickmann, A.; Weber, G.; Grosse, J.; Infanger, M.; Eilles, C.; Grimm, D. Interaction of proteins identified in human thyroid cells. Int. J. Mol. Sci. 2013, 14, 1164–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Wang, C.; Sun, S.; Zhang, C.; Lu, D.; Chen, Q.; Long, M. Microgravity-Induced Alterations of Inflammation-Related Mechanotransduction in Endothelial Cells on Board SJ-10 Satellite. Front. Physiol. 2018, 9, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorchenko, O.; Stiefelhagen, M.; Peer-Zada, A.A.; Barthel, R.; Mayer, P.; Eckei, L.; Breuer, A.; Crispatzu, G.; Rosen, N.; Landwehr, T.; et al. CD44 regulates the apoptotic response and promotes disease development in chronic lymphocytic leukemia. Blood 2013, 121, 4126–4136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, M.L.; Reynolds, J.L.; Cubano, L.A.; Hatton, J.P.; Lawless, B.D.; Piepmeier, E.H. Spaceflight alters microtubules and increases apoptosis in human lymphocytes (Jurkat). FASEB J. 1998, 12, 1007–1018. [Google Scholar] [CrossRef]










| Factor | Primer Name | Sequence 5’–3’ |
|---|---|---|
| 18S rRNA | 18s-F | GGAGCCTGCGGCTTAATTT |
| 18s-R | CAACTAAGAACGGCCATGCA | |
| Annexin A1; ANXA1 | ANXA1-F | GCCAAAGACATAACCTCAGACACAT |
| ANXA1-R | GAATCAGCCAAGTCTTCATTCACA | |
| Annexin A2; ANXA2 | ANXA2-F | GGTACAAGAGTTACAGCCCTTATGACA |
| ANXA2-R | CATGGAGTCATACAGCCGATCA | |
| BCL2 Associated X, Apoptosis Regulator; BAX | Bax-F | GTCAGCTGCCACTCGGAAA |
| Bax-R | AGTAACATGGAGCTGCAGAGGAT | |
| B-cell lymphoma 2; BCL2 | Bcl2-F | TCAGAGACAGCCAGGAGAAATCA |
| Bcl2-R | CCTGTGGATGACTGAGTACCTGAA | |
| Caspase 3; CASP3 | Casp3-F | CTCCAACATCGACTGTGAGAAGTT |
| Casp3-R | GCGCCAGCTCCAGCAA | |
| CD44 | CD44-F | ACCCTCCCCTCATTCACCAT |
| CD44-R | GTTGTACTACTAGGAGTTGCCTGGATT | |
| Extracellular signal-regulated kinases 1; ERK1 | ERK1-F | ACCTGCGACCTTAAGATTTGTGA |
| ERK1-R | AGCCACATACTCCGTCAGGAA | |
| Extracellular signal-regulated kinases 2; ERK2 | ERK2-F | TTCCAACCTGCTGCTCAACA |
| ERK2-R | TCTGTCAGGAACCCTGTGTGAT | |
| Focal adhesion kinase 1 (Protein-tyrosine kinase 2); pan-FAK1 | FAK1-F | TGTGGGTAAACCAGATCCTGC |
| FAK1-R | CTGAAGCTTGACACCCTCGT | |
| Intercellular adhesion molecule 1; ICAM1 | ICAM1-F | CGGCTGACGTGTGCAGTAAT |
| ICAM1-R | CTTCTGAGACCTCTGGCTTCGT | |
| Mitogen-activated protein kinase 8 (MAPK8) (JNK1-a2); MAPK8/JNK1 | JNK1-F | TCTCCTTTAGGTGCAGCAGTG |
| JNK1-R | CAGAGGCCAAAGTCGGATCT | |
| NF-kappa-B transcription complex P105/P50; NFKB1 | NFkB1-F | CTTAGGAGGGAGAGCCCAC |
| NFkB1-R | TGAAACATTTGTTCAGGCCTTC | |
| NF-kappa-B transcription complex P100/P52; NFKB2 | NFkB2-F | GTACAAAGATACGCGGACCC |
| NFkB2-R | CCAGACCTGGGTTGTAGCA | |
| NF-kappa-B transcription complex P65; NFKB P65 | NFkB P65-F | CGCTTCTTCACACACTGGATTC |
| NFkB P65-R | ACTGCCGGGATGGCTTCT | |
| NF-kappa-B essential modulator (NEMO); IKBKG | IkBKG-F | AACTGGGACTTTCTCGGAGC |
| IkBKG-R | GGCAAGGGCTGTCAGCAG | |
| NF-kappa-B inhibitor alpha; NFKBIA | NFkBIa-F | AATGCTCAGGAGCCCTGTAAT |
| NFkBIa-R | CTGTTGACATCAGCCCCACA | |
| NF-kappa-B inhibitor beta; NFKBIB | NFkBIb-F | CCCGGAGGACCTGGGTT |
| NFkBIb-R | GCAGTGCCGTGTCCCC | |
| NF-kappa-B inhibitor epsilon; NFKBIE | NFkBIe-F | TGGGCATCTCATCCACTCTG |
| NFkBIe-R | ACAAGGGATTCCTCAGTCAGGT | |
| Protein kinase C alpha type; PRKCA | PRKCA-F | TGGGTCACTGCTCTATGGACTTATC |
| PRKCA-R | CGCCCCCTCTTCTCAGTGT | |
| TATA-box binding protein; TBP | TBP-F | GTGACCCAGCATCACTGTTTC |
| TBP-R | GCAAACCAGAAACCCTTGCG | |
| Raf-1 Proto-Oncogene, Serine/Threonine Kinase; Raf1 | Raf1-F | GGGAGCTTGGAAGACGATCAG |
| Raf1-R | ACACGGATAGTGTTGCTTGTC | |
| Osteopontin (OPN); SPP1 | SPP1-F | CGAGGTGATAGTGTGGTTTATGGA |
| SPP1-R | CGTCTGTAGCATCAGGGTACTG | |
| Vascular cell adhesion protein 1; VCAM1 | VCAM1-F | CATGGAATTCGAACCCAAACA |
| VCAM1-R | GGCTGACCAAGACGGTTGTATC |
| Antibody | kDa | Dilution | Company | Source |
|---|---|---|---|---|
| Annexin 2 | 38 | 1:1000 | Abcam #ab41802 | Rb |
| NFkBp65 | 65 | 1:1000 | Thermo Fisher #PA1-186 | Rb |
| IKBKG | 38 | 1:500 | Origene #TA812460 | MS |
| IkBα/NFKBIA | 36 | 1:1000 | Invitrogen #MA5-15132 | MS |
| Cofilin-1 | 20 | 1:2000 | Abcam #ab 42824 | Rb |
| CD44 | 80 | 1:500 | CST#5640 | MS |
| VCAM1 | 110 | 1:500 | Sc80431 | MS |
| Casp 3 | 35 | 1:800 | CST#9662 | Rb |
| ICAM1 | 90 | 1:500 | CST#4915 | Rb |
| Osteopontin | 44 | 1:1000 | SAB4200018 | MS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nassef, M.Z.; Kopp, S.; Melnik, D.; Corydon, T.J.; Sahana, J.; Krüger, M.; Wehland, M.; Bauer, T.J.; Liemersdorf, C.; Hemmersbach, R.; et al. Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 5730. https://doi.org/10.3390/ijms20225730
Nassef MZ, Kopp S, Melnik D, Corydon TJ, Sahana J, Krüger M, Wehland M, Bauer TJ, Liemersdorf C, Hemmersbach R, et al. Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells. International Journal of Molecular Sciences. 2019; 20(22):5730. https://doi.org/10.3390/ijms20225730
Chicago/Turabian StyleNassef, Mohamed Zakaria, Sascha Kopp, Daniela Melnik, Thomas J. Corydon, Jayashree Sahana, Marcus Krüger, Markus Wehland, Thomas J. Bauer, Christian Liemersdorf, Ruth Hemmersbach, and et al. 2019. "Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells" International Journal of Molecular Sciences 20, no. 22: 5730. https://doi.org/10.3390/ijms20225730
APA StyleNassef, M. Z., Kopp, S., Melnik, D., Corydon, T. J., Sahana, J., Krüger, M., Wehland, M., Bauer, T. J., Liemersdorf, C., Hemmersbach, R., Infanger, M., & Grimm, D. (2019). Short-Term Microgravity Influences Cell Adhesion in Human Breast Cancer Cells. International Journal of Molecular Sciences, 20(22), 5730. https://doi.org/10.3390/ijms20225730