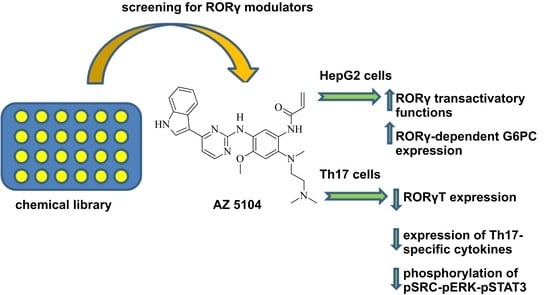
The Dichotomous Nature of AZ5104 (an EGFR Inhibitor) Towards RORγ and RORγT
Abstract
:1. Introduction
2. Results
2.1. Identification of AZ5104 as a RORγ Activator
2.2. Molecular Docking Analysis
2.3. AZ5104 Inhibits Th17-Specific Genes Expression
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Screening of the Chemical Library
4.3. Cell Viability
4.4. Vectors, Transfection and Luciferase Assay
4.5. Naive CD4+ T Cell Isolation and Differentiation into Th17 Cells
4.6. Real-Time RT-PCR
4.7. siRNA Methodology
4.8. Chromatin Immunoprecipitation (ChIP)
4.9. Glucose 6-Phosphate Assay
4.10. Detection and Quantification of IL-17 (ELISA)
4.11. Western Blotting
4.12. Docking Simulations
4.13. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACT | adoptive cell therapy |
| AHR | aryl hydrocarbon receptor |
| BATF | basic leucine zipper ATF-like transcription factor |
| CD4+ | CD4+ helper T cells |
| CSF2 | colony stimulating factor 2 |
| DBD | DNA-binding domain |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| ERK | extracellular regulated MAP kinase |
| FOXP3 | forkhead box protein P3 |
| G6PC | glucose-6-phosphatase catalytic subunit |
| GATA3 | GATA binding protein 3 |
| GM-CSF2 | colony stimulating factor 2 (granulocyte-macrophage) |
| HMBS | hydroxymethylbilane synthase |
| HPRT1 | hypoxanthine phosphoribosyltransferase 1 |
| IL9 | interleukin 9 |
| IL17A | interleukin 17A |
| IL17F | interleukin 17F |
| IL-21 | interleukin 21 |
| IL-22 | interleukin 22 |
| IL-23 | interleukin 23A |
| IRF4 | interferon regulatory factor 4 |
| LBD | ligand binding domain |
| NR1F3 | nuclear receptor subfamily 1 group F member 3 |
| PPAR | peroxisome proliferator-activated receptor |
| RORC | RAR-related orphan receptor gamma |
| RORγ | nuclear receptor ROR-gamma isoform 1 |
| RORγT | nuclear receptor ROR-gamma isoform 2 |
| RPL13A | ribosomal protein L13a |
| STAT3 | signal transducer and activator of transcription 3 |
| SRC | SRC proto-oncogene, non-receptor tyrosine kinase |
| t-bet | cell-specific T-box transcription factor |
| TGF-β | transforming growth factor beta |
| Th17 | T-helper 17 cells |
References
- Harris, K.M.; Ramachandran, G.; Basu, S.; Rollins, S.; Mann, D.; Cross, A.S. The il-23/th17 axis is involved in the adaptive immune response to bacillus anthracis in humans. Eur. J. Immunol. 2014, 44, 752–762. [Google Scholar] [CrossRef]
- Lin, L.; Ibrahim, A.S.; Xu, X.; Farber, J.M.; Avanesian, V.; Baquir, B.; Fu, Y.; French, S.W.; Edwards, J.E., Jr.; Spellberg, B. Th1-th17 cells mediate protective adaptive immunity against staphylococcus aureus and candida albicans infection in mice. PLoS Pathog. 2009, 5, e1000703. [Google Scholar] [CrossRef]
- Huang, W.; Na, L.; Fidel, P.L.; Schwarzenberger, P. Requirement of interleukin-17a for systemic anti-candida albicans host defense in mice. J. Infect. Dis. 2004, 190, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.S.; Chew, G.Y.; Simpson, N.; Priyadarshi, A.; Wong, M.; Grimbacher, B.; Fulcher, D.A.; Tangye, S.G.; Cook, M.C. Deficiency of th17 cells in hyper ige syndrome due to mutations in stat3. J. Exp. Med. 2008, 205, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, K.; Foerster, S.; Rombold, S.; Seidl, H.P.; Behrendt, H.; Hofmann, H.; Ring, J.; Traidl-Hoffmann, C. Patients with chronic mucocutaneous candidiasis exhibit reduced production of th17-associated cytokines il-17 and il-22. J. Investig. Dermatol. 2008, 128, 2640–2645. [Google Scholar] [CrossRef]
- Hirota, K.; Hashimoto, M.; Yoshitomi, H.; Tanaka, S.; Nomura, T.; Yamaguchi, T.; Iwakura, Y.; Sakaguchi, N.; Sakaguchi, S. T cell self-reactivity forms a cytokine milieu for spontaneous development of il-17+ th cells that cause autoimmune arthritis. J. Exp. Med. 2007, 204, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, X.; Liu, Z.; Yue, Q.; Liu, H. Expression of th17 cytokines in skin lesions of patients with psoriasis. J. Huazhong Univ. Sci. Technol. Med. Sci. 2007, 27, 330–332. [Google Scholar] [CrossRef] [PubMed]
- Kebir, H.; Kreymborg, K.; Ifergan, I.; Dodelet-Devillers, A.; Cayrol, R.; Bernard, M.; Giuliani, F.; Arbour, N.; Becher, B.; Prat, A. Human th17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 2007, 13, 1173–1175. [Google Scholar] [CrossRef]
- Jandus, C.; Bioley, G.; Rivals, J.P.; Dudler, J.; Speiser, D.; Romero, P. Increased numbers of circulating polyfunctional th17 memory cells in patients with seronegative spondylarthritides. Arthritis Rheum. 2008, 58, 2307–2317. [Google Scholar] [CrossRef]
- Annunziato, F.; Cosmi, L.; Santarlasci, V.; Maggi, L.; Liotta, F.; Mazzinghi, B.; Parente, E.; Fili, L.; Ferri, S.; Frosali, F.; et al. Phenotypic and functional features of human th17 cells. J. Exp. Med. 2007, 204, 1849–1861. [Google Scholar] [CrossRef]
- Yang, X.O.; Panopoulos, A.D.; Nurieva, R.; Chang, S.H.; Wang, D.; Watowich, S.S.; Dong, C. Stat3 regulates cytokine-mediated generation of inflammatory helper t cells. J. Biol. Chem. 2007, 282, 9358–9363. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Ogura, H.; Ueda, N.; Tsuruoka, M.; Kitabayashi, C.; Tsuji, F.; Aono, H.; Ishihara, K.; Huseby, E.; Betz, U.A.; et al. Il-6-gp130-stat3 in t cells directs the development of il-17+ th with a minimum effect on that of treg in the steady state. Int. Immunol. 2007, 19, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Bettelli, E.; Korn, T.; Kuchroo, V.K. Th17: The third member of the effector t cell trilogy. Curr. Opin. Immunol. 2007, 19, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.J.; Grosso, J.F.; Yen, H.R.; Xin, H.; Kortylewski, M.; Albesiano, E.; Hipkiss, E.L.; Getnet, D.; Goldberg, M.V.; Maris, C.H.; et al. Cutting edge: An in vivo requirement for stat3 signaling in th17 development and th17-dependent autoimmunity. J. Immunol. 2007, 179, 4313–4317. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Laurence, A.; Elias, K.M.; O’Shea, J.J. Il-21 is produced by th17 cells and drives il-17 production in a stat3-dependent manner. J. Biol. Chem. 2007, 282, 34605–34610. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Brustle, A.; Reinhard, K.; Guralnik, A.; Walter, G.; Mahiny, A.; von Low, E.; Lohoff, M. Irf4 is essential for il-21-mediated induction, amplification, and stabilization of the th17 phenotype. Proc. Natl. Acad. Sci. USA 2008, 105, 20846–20851. [Google Scholar] [CrossRef]
- Schraml, B.U.; Hildner, K.; Ise, W.; Lee, W.L.; Smith, W.A.; Solomon, B.; Sahota, G.; Sim, J.; Mukasa, R.; Cemerski, S.; et al. The ap-1 transcription factor batf controls t(h)17 differentiation. Nature 2009, 460, 405–409. [Google Scholar] [CrossRef]
- Martinez, G.J.; Dong, C. Batf: Bringing (in) another th17-regulating factor. J. Mol. Cell Biol. 2009, 1, 66–68. [Google Scholar] [CrossRef]
- Ivanov, I.I.; McKenzie, B.S.; Zhou, L.; Tadokoro, C.E.; Lepelley, A.; Lafaille, J.J.; Cua, D.J.; Littman, D.R. The orphan nuclear receptor rorgammat directs the differentiation program of proinflammatory il-17+ t helper cells. Cell 2006, 126, 1121–1133. [Google Scholar] [CrossRef]
- Szabo, S.J.; Kim, S.T.; Costa, G.L.; Zhang, X.; Fathman, C.G.; Glimcher, L.H. A novel transcription factor, t-bet, directs th1 lineage commitment. Cell 2000, 100, 655–669. [Google Scholar] [CrossRef]
- Zheng, W.; Flavell, R.A. The transcription factor gata-3 is necessary and sufficient for th2 cytokine gene expression in cd4 t cells. Cell 1997, 89, 587–596. [Google Scholar] [CrossRef]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral cd4+cd25- naive t cells to cd4+cd25+ regulatory t cells by tgf-beta induction of transcription factor foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Atkins, G.B.; Hu, X.; Guenther, M.G.; Rachez, C.; Freedman, L.P.; Lazar, M.A. Coactivators for the orphan nuclear receptor roralpha. Mol. Endocrinol. 1999, 13, 1550–1557. [Google Scholar] [PubMed]
- Xie, H.; Sadim, M.S.; Sun, Z. Rorgammat recruits steroid receptor coactivators to ensure thymocyte survival. J. Immunol. 2005, 175, 3800–3809. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Escriva Garcia, H.; Laudet, V. The nuclear receptor superfamily. J. Cell Sci. 2003, 116, 585–586. [Google Scholar] [CrossRef]
- Medvedev, A.; Chistokhina, A.; Hirose, T.; Jetten, A.M. Genomic structure and chromosomal mapping of the nuclear orphan receptor ror gamma (rorc) gene. Genomics 1997, 46, 93–102. [Google Scholar] [CrossRef]
- Villey, I.; de Chasseval, R.; de Villartay, J.P. Rorgammat, a thymus-specific isoform of the orphan nuclear receptor rorgamma/tor, is up-regulated by signaling through the pre-t cell receptor and binds to the tea promoter. Eur. J. Immunol. 1999, 29, 4072–4080. [Google Scholar] [CrossRef]
- Ratajewski, M.; Walczak-Drzewiecka, A.; Salkowska, A.; Dastych, J. Upstream stimulating factors regulate the expression of rorgammat in human lymphocytes. J. Immunol. 2012, 189, 3034–3042. [Google Scholar] [CrossRef]
- Crome, S.Q.; Wang, A.Y.; Kang, C.Y.; Levings, M.K. The role of retinoic acid-related orphan receptor variant 2 and il-17 in the development and function of human cd4+ t cells. Eur. J. Immunol. 2009, 39, 1480–1493. [Google Scholar] [CrossRef]
- He, Y.W.; Deftos, M.L.; Ojala, E.W.; Bevan, M.J. Rorgamma t, a novel isoform of an orphan receptor, negatively regulates fas ligand expression and il-2 production in t cells. Immunity 1998, 9, 797–806. [Google Scholar] [CrossRef]
- Kang, H.S.; Angers, M.; Beak, J.Y.; Wu, X.; Gimble, J.M.; Wada, T.; Xie, W.; Collins, J.B.; Grissom, S.F.; Jetten, A.M. Gene expression profiling reveals a regulatory role for ror alpha and ror gamma in phase i and phase ii metabolism. Physiol. Genom. 2007, 31, 281–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jetten, A.M. Retinoid-related orphan receptors (rors): Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 2009, 7, e003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, Y.; Jothi, R.; Birault, V.; Jetten, A.M. Rorgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012, 40, 8519–8535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, J.R.; Littman, D.R. Small molecule inhibitors of rorgammat: Targeting th17 cells and other applications. Eur. J. Immunol. 2012, 42, 2232–2237. [Google Scholar] [CrossRef] [Green Version]
- Qiu, R.; Wang, Y. Retinoic acid receptor-related orphan receptor gammat (rorgammat) agonists as potential small molecule therapeutics for cancer immunotherapy. J. Med. Chem. 2018, 61, 5794–5804. [Google Scholar] [CrossRef]
- Dal Pra, M.; Carta, D.; Szabadkai, G.; Suman, M.; Frion-Herrera, Y.; Paccagnella, N.; Castellani, G.; De Martin, S.; Ferlin, M.G. Targeting rors nuclear receptors by novel synthetic steroidal inverse agonists for autoimmune disorders. Bioorg. Med. Chem. 2018, 26, 1686–1704. [Google Scholar] [CrossRef]
- Karas, K.; Salkowska, A.; Walczak-Drzewiecka, A.; Ryba, K.; Dastych, J.; Bachorz, R.A.; Ratajewski, M. The cardenolides strophanthidin, digoxigenin and dihydroouabain act as activators of the human rorgamma/rorgammat receptors. Toxicol. Lett. 2018, 295, 314–324. [Google Scholar] [CrossRef]
- Wang, Y.; Kumar, N.; Solt, L.A.; Richardson, T.I.; Helvering, L.M.; Crumbley, C.; Garcia-Ordonez, R.D.; Stayrook, K.R.; Zhang, X.; Novick, S.; et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. J. Biol. Chem. 2010, 285, 5013–5025. [Google Scholar] [CrossRef] [Green Version]
- Takeda, Y.; Kang, H.S.; Freudenberg, J.; DeGraff, L.M.; Jothi, R.; Jetten, A.M. Retinoic acid-related orphan receptor gamma (rorgamma): A novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. 2014, 10, e1004331. [Google Scholar] [CrossRef]
- Yates, J.W.; Ashton, S.; Cross, D.; Mellor, M.J.; Powell, S.J.; Ballard, P. Irreversible inhibition of egfr: Modeling the combined pharmacokinetic-pharmacodynamic relationship of osimertinib and its active metabolite az5104. Mol. Cancer Ther. 2016, 15, 2378–2387. [Google Scholar] [CrossRef]
- Croswell, J.M.; Kramer, B.S.; Kreimer, A.R.; Prorok, P.C.; Xu, J.L.; Baker, S.G.; Fagerstrom, R.; Riley, T.L.; Clapp, J.D.; Berg, C.D.; et al. Cumulative incidence of false-positive results in repeated, multimodal cancer screening. Ann. Fam. Med. 2009, 7, 212–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copik, A.J.; Webb, M.S.; Miller, A.L.; Wang, Y.; Kumar, R.; Thompson, E.B. Activation function 1 of glucocorticoid receptor binds tata-binding protein in vitro and in vivo. Mol. Endocrinol. 2006, 20, 1218–1230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, E.C.; Noelle, R.J. Interleukin-9 as a t helper type 17 cytokine. Immunology 2010, 131, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.C.; Tan, X.Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (il)-22 and il-17 are coexpressed by th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef]
- Sonderegger, I.; Iezzi, G.; Maier, R.; Schmitz, N.; Kurrer, M.; Kopf, M. Gm-csf mediates autoimmunity by enhancing il-6-dependent th17 cell development and survival. J. Exp. Med. 2008, 205, 2281–2294. [Google Scholar] [CrossRef]
- Stritesky, G.L.; Yeh, N.; Kaplan, M.H. Il-23 promotes maintenance but not commitment to the th17 lineage. J. Immunol. 2008, 181, 5948–5955. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Cheng, F.; Zhao, Z. Tissue-specific signaling networks rewired by major somatic mutations in human cancer revealed by proteome-wide discovery. Cancer Res. 2017, 77, 2810–2821. [Google Scholar] [CrossRef]
- Zhang, Z.; Burch, P.E.; Cooney, A.J.; Lanz, R.B.; Pereira, F.A.; Wu, J.; Gibbs, R.A.; Weinstock, G.; Wheeler, D.A. Genomic analysis of the nuclear receptor family: New insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004, 14, 580–590. [Google Scholar] [CrossRef] [Green Version]
- Parrado, A.; Despouy, G.; Kraiba, R.; Le Pogam, C.; Dupas, S.; Choquette, M.; Robledo, M.; Larghero, J.; Bui, H.; Le Gall, I.; et al. Retinoic acid receptor alpha1 variants, raralpha1deltab and raralpha1deltabc, define a new class of nuclear receptor isoforms. Nucleic Acids Res. 2001, 29, 4901–4908. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Cidlowski, J.A. The human glucocorticoid receptor: One gene, multiple proteins and diverse responses. Steroids 2005, 70, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Sever, R.; Glass, C.K. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013, 5, a016709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Chandra, V.; Rastinejad, F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu. Rev. Physiol. 2010, 72, 247–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moseson, D.L.; Sasaki, G.H.; Kraybill, W.G.; Leung, B.S.; Davenport, C.E.; Fletcher, W.S. The use of antiestrogens tamoxifen and nafoxidine in the treatment of human breast cancer in correlation with estrogen receptor values. A phase ii study. Cancer 1978, 41, 797–802. [Google Scholar] [CrossRef]
- Scher, H.I.; Beer, T.M.; Higano, C.S.; Anand, A.; Taplin, M.E.; Efstathiou, E.; Rathkopf, D.; Shelkey, J.; Yu, E.Y.; Alumkal, J.; et al. Antitumour activity of mdv3100 in castration-resistant prostate cancer: A phase 1-2 study. Lancet 2010, 375, 1437–1446. [Google Scholar] [CrossRef] [Green Version]
- Seeman, E. Raloxifene. J. Bone Miner. Metab. 2001, 19, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.J. Glucocorticoid action and the development of selective glucocorticoid receptor ligands. Biotechnol. Annu. Rev. 2006, 12, 269–300. [Google Scholar] [PubMed]
- Konig, B.; Koch, A.; Spielmann, J.; Hilgenfeld, C.; Stangl, G.I.; Eder, K. Activation of pparalpha lowers synthesis and concentration of cholesterol by reduction of nuclear srebp-2. Biochem. Pharmacol. 2007, 73, 574–585. [Google Scholar] [CrossRef]
- Rodney, G.; Uhlendorf, P.; Maxwell, R.E. The hypolipidaemic effect of gemfibrozil (ci-719) in laboratory animals. Proc. R. Soc. Med. 1976, 69, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Bailey, S.R.; Nelson, M.H.; Himes, R.A.; Li, Z.; Mehrotra, S.; Paulos, C.M. Th17 cells in cancer: The ultimate identity crisis. Front. Immunol. 2014, 5, 276. [Google Scholar] [CrossRef] [Green Version]
- Muranski, P.; Boni, A.; Antony, P.A.; Cassard, L.; Irvine, K.R.; Kaiser, A.; Paulos, C.M.; Palmer, D.C.; Touloukian, C.E.; Ptak, K.; et al. Tumor-specific th17-polarized cells eradicate large established melanoma. Blood 2008, 112, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Canderan, G.; Dellabona, P. T helper 17 t cells do good for cancer immunotherapy. Immunotherapy 2010, 2, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.S.; Nelson, M.H.; Majchrzak, K.; Bailey, S.R.; Rohrer, B.; Kaiser, A.D.; Atkinson, C.; Gattinoni, L.; Paulos, C.M. Th17 cells are refractory to senescence and retain robust antitumor activity after long-term ex vivo expansion. JCI Insight 2017, 2, e90772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, O.K.; Schaefer, T.S.; Nathans, D. In vitro activation of stat3 by epidermal growth factor receptor kinase. Proc. Natl. Acad. Sci. USA 1996, 93, 13704–13708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aznar, S.; Valeron, P.F.; del Rincon, S.V.; Perez, L.F.; Perona, R.; Lacal, J.C. Simultaneous tyrosine and serine phosphorylation of stat3 transcription factor is involved in rho a gtpase oncogenic transformation. Mol. Biol. Cell 2001, 12, 3282–3294. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.P.; Cao, X. Serine phosphorylation and negative regulation of stat3 by jnk. J. Biol. Chem. 1999, 274, 31055–31061. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.P.; Cao, X. Regulation of stat3 activation by mek kinase 1. J. Biol. Chem. 2001, 276, 21004–21011. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.J.; Hwang, Y.J.; Yun, M.O.; Kim, J.H.; Oh, G.S.; Park, J.H. Indoxyl 3-sulfate stimulates th17 differentiation enhancing phosphorylation of c-src and stat3 to worsen experimental autoimmune encephalomyelitis. Toxicol. Lett. 2013, 220, 109–117. [Google Scholar] [CrossRef]
- Liu, H.; Yao, S.; Dann, S.M.; Qin, H.; Elson, C.O.; Cong, Y. Erk differentially regulates th17- and treg-cell development and contributes to the pathogenesis of colitis. Eur. J. Immunol. 2013, 43, 1716–1726. [Google Scholar] [CrossRef]
- El-Hashim, A.Z.; Khajah, M.A.; Renno, W.M.; Babyson, R.S.; Uddin, M.; Benter, I.F.; Ezeamuzie, C.; Akhtar, S. Src-dependent egfr transactivation regulates lung inflammation via downstream signaling involving erk1/2, pi3kdelta/akt and nfkappab induction in a murine asthma model. Sci. Rep. 2017, 7, 9919. [Google Scholar] [CrossRef]
- Bullens, D.M.; Truyen, E.; Coteur, L.; Dilissen, E.; Hellings, P.W.; Dupont, L.J.; Ceuppens, J.L. Il-17 mrna in sputum of asthmatic patients: Linking t cell driven inflammation and granulocytic influx? Respir. Res. 2006, 7, 135. [Google Scholar] [CrossRef] [Green Version]
- Al-Ramli, W.; Al Samri, M.; Hamid, Q. Th-17 cell-related cytokines’ potential role in the pathogenesis of severe asthma. J. Asthma 2008, 45, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Eilers, R.E., Jr.; Gandhi, M.; Patel, J.D.; Mulcahy, M.F.; Agulnik, M.; Hensing, T.; Lacouture, M.E. Dermatologic infections in cancer patients treated with epidermal growth factor receptor inhibitor therapy. J. Natl. Cancer Inst. 2010, 102, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.Y.; Lee, Y.J.; Kim, J.M.; Kang, H.J.; Cho, S.H.; Chang, S.E. Epidermal growth factor relieves inflammatory signals in staphylococcus aureus-treated human epidermal keratinocytes and atopic dermatitis-like skin lesions in nc/nga mice. BioMed Res. Int. 2018, 2018, 9439182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finlay, M.R.; Anderton, M.; Ashton, S.; Ballard, P.; Bethel, P.A.; Box, M.R.; Bradbury, R.H.; Brown, S.J.; Butterworth, S.; Campbell, A.; et al. Discovery of a potent and selective egfr inhibitor (azd9291) of both sensitizing and t790m resistance mutations that spares the wild type form of the receptor. J. Med. Chem. 2014, 57, 8249–8267. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.A.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.; Ward, R.A.; Mellor, M.J.; et al. Azd9291, an irreversible egfr tki, overcomes t790m-mediated resistance to egfr inhibitors in lung cancer. Cancer Discov. 2014, 4, 1046–1061. [Google Scholar] [CrossRef] [PubMed]
- Carlisle, J.W.; Ramalingam, S.S. Role of osimertinib in the treatment of egfr-mutation positive non-small-cell lung cancer. Future Oncol. 2019, 15, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Takagi, Y.; Okubo, H.; Yamaguchi, S.; Kikkawa, Y.; Hashimoto, I.; Kaburagi, T.; Miura, M.; Satoh, H.; Hizawa, N. Plasma concentration of osimertinib in a non-small cell lung cancer patient with chronic renal failure undergoing hemodialysis. Lung Cancer 2017, 112, 225–226. [Google Scholar] [CrossRef]
- Rood, J.J.M.; van Haren, M.J.; Beijnen, J.H.; Sparidans, R.W. Bioanalysis of egfrm inhibitor osimertinib, and its glutathione cycle- and desmethyl metabolites by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2019, 177, 112871. [Google Scholar] [CrossRef]
- Salkowska, A.; Karas, K.; Walczak-Drzewiecka, A.; Dastych, J.; Ratajewski, M. Differentiation stage-specific effect of histone deacetylase inhibitors on the expression of rorgammat in human lymphocytes. J. Leukoc. Biol. 2017, 102, 1487–1495. [Google Scholar] [CrossRef]
- Karas, K.; Salkowska, A.; Sobalska-Kwapis, M.; Walczak-Drzewiecka, A.; Strapagiel, D.; Dastych, J.; Bachorz, R.A.; Ratajewski, M. Digoxin, an overlooked agonist of rorgamma/rorgammat. Front. Pharmacol. 2019, 9, 1460. [Google Scholar] [CrossRef]
- Repetto, G.; del Peso, A.; Zurita, J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 2008, 3, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Stehlin-Gaon, C.; Willmann, D.; Zeyer, D.; Sanglier, S.; Van Dorsselaer, A.; Renaud, J.P.; Moras, D.; Schule, R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ror beta. Nat. Struct. Biol. 2003, 10, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Solt, L.A.; Conkright, J.J.; Wang, Y.; Istrate, M.A.; Busby, S.A.; Garcia-Ordonez, R.D.; Burris, T.P.; Griffin, P.R. The benzenesulfoamide t0901317 [n-(2,2,2-trifluoroethyl)-n-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethy l]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol. Pharmacol. 2010, 77, 228–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, N.J.; Boniface, K.; Chan, J.R.; McKenzie, B.S.; Blumenschein, W.M.; Mattson, J.D.; Basham, B.; Smith, K.; Chen, T.; Morel, F.; et al. Development, cytokine profile and function of human interleukin 17-producing helper t cells. Nat. Immunol. 2007, 8, 950–957. [Google Scholar] [CrossRef]
- Ratajewski, M.; Walczak-Drzewiecka, A.; Gorzkiewicz, M.; Salkowska, A.; Dastych, J. Expression of human gene coding rorgammat receptor depends on the sp2 transcription factor. J. Leukoc. Biol. 2016, 100, 1213–1223. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. Pubchem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Hawkins, P.C.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer generation with omega: Algorithm and validation using high quality structures from the protein databank and cambridge structural database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef]
- Santa Fe, N.M. Omega 3.1.1.2: Openeye Scientific Software. Available online: http://www.eyesopen.com (accessed on 31 May 2019).
- Fujita-Sato, S.; Ito, S.; Isobe, T.; Ohyama, T.; Wakabayashi, K.; Morishita, K.; Ando, O.; Isono, F. Structural basis of digoxin that antagonizes rorgamma t receptor activity and suppresses th17 cell differentiation and interleukin (il)-17 production. J. Biol. Chem. 2011, 286, 31409–31417. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Martynowski, D.; Zheng, S.; Wada, T.; Xie, W.; Li, Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor rorgamma. Mol. Endocrinol. 2010, 24, 923–929. [Google Scholar] [CrossRef]
- Ahn, M.J.; Tsai, C.M.; Shepherd, F.A.; Bazhenova, L.; Sequist, L.V.; Hida, T.; Yang, J.C.H.; Ramalingam, S.S.; Mitsudomi, T.; Janne, P.A.; et al. Osimertinib in patients with t790m mutation-positive, advanced non-small cell lung cancer: Long-term follow-up from a pooled analysis of 2 phase 2 studies. Cancer 2019, 125, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Mezquita, L.; Varga, A.; Planchard, D. Safety of osimertinib in egfr-mutated non-small cell lung cancer. Expert Opin. Drug Saf. 2018, 17, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.M.; Huang, J.A.; Chen, Y.B. Fatal interstitial lung disease associated with azd9291. J. Cancer Res. Ther. 2018, 14, S1227–S1229. [Google Scholar] [PubMed]








| Host Domain | Estimated Free Energy of Binding (kcal/mol) | Number of Items in Cluster | |
|---|---|---|---|
| Lowest | Mean | ||
| 3L01 | −9.53 | −8.83 | 3 |
| 3B0W_A | −8.90 | −8.90 | 1 |
| 3B0W_B | −9.04 | −9.04 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaś, K.; Sałkowska, A.; Karwaciak, I.; Walczak-Drzewiecka, A.; Dastych, J.; Bachorz, R.A.; Ratajewski, M. The Dichotomous Nature of AZ5104 (an EGFR Inhibitor) Towards RORγ and RORγT. Int. J. Mol. Sci. 2019, 20, 5780. https://doi.org/10.3390/ijms20225780
Karaś K, Sałkowska A, Karwaciak I, Walczak-Drzewiecka A, Dastych J, Bachorz RA, Ratajewski M. The Dichotomous Nature of AZ5104 (an EGFR Inhibitor) Towards RORγ and RORγT. International Journal of Molecular Sciences. 2019; 20(22):5780. https://doi.org/10.3390/ijms20225780
Chicago/Turabian StyleKaraś, Kaja, Anna Sałkowska, Iwona Karwaciak, Aurelia Walczak-Drzewiecka, Jarosław Dastych, Rafał A. Bachorz, and Marcin Ratajewski. 2019. "The Dichotomous Nature of AZ5104 (an EGFR Inhibitor) Towards RORγ and RORγT" International Journal of Molecular Sciences 20, no. 22: 5780. https://doi.org/10.3390/ijms20225780
APA StyleKaraś, K., Sałkowska, A., Karwaciak, I., Walczak-Drzewiecka, A., Dastych, J., Bachorz, R. A., & Ratajewski, M. (2019). The Dichotomous Nature of AZ5104 (an EGFR Inhibitor) Towards RORγ and RORγT. International Journal of Molecular Sciences, 20(22), 5780. https://doi.org/10.3390/ijms20225780