Daphnetin: A Novel Anti-Helicobacter pylori Agent
Abstract
:1. Introduction
2. Results
2.1. Antibacterial Activity of Daphnetin Against H. pylori Strains
2.2. Effect of Daphnetin on H. pylori Morphology
2.3. Daphnetin-Induced Membrane Changes
2.4. Daphnetin Caused DNA Damage
2.5. Daphnetin Decreased H. pylori Adherence to Immortalized Human Gastric Epithelial Cell Line (GES-1) and Inhibited Colonization-Associated Gene Expression
2.6. The Cytotoxic Effect of Daphnetin on GES-1
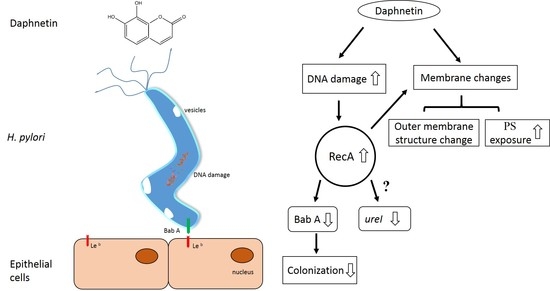
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Materials
4.2. Cell Cultures
4.3. MICs of Daphnetin, Metronidazole, and Clarithromycin on H. pylori Strains
4.4. H. pylori Morphology Analyses by SEM and TEM
4.5. Detection of Membrane Changes
4.6. Detection of DNA Damage
4.7. RNA Isolation and Quantitative Real-Time PCR
4.8. Membrane Preparation and Proteomics by Liquid Chromatography–Mass Spectrometry/Mass Spectrometry Analyses
4.9. H. pylori Adhesion Assays
4.10. Cell Cytotoxicity Assays
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Warren, J.R.; Marshall, B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983, 321, 1273–1275. [Google Scholar]
- Zali, M.R. Facing resistance of H. pylori infection. Gastroenterol. Hepatol. Bed Bench 2011, 4, 3–11. [Google Scholar] [PubMed]
- Walker, M.M.; Crabtree, J.E. Helicobacter pylori infection and the pathogenesis of duodenal ulceration. Ann. N. Y. Acad. Sci. 1998, 859, 96–111. [Google Scholar] [CrossRef] [PubMed]
- McNulty, C.; Owen, R.; Tompkins, D.; Hawtin, P.; McColl, K.; Price, A.; Smith, G.; Teare, L. Helicobacter pylori susceptibility testing by disc diffusion. J. Antimicrob. Chemother. 2002, 49, 601–609. [Google Scholar] [CrossRef]
- Chey, W.D.; I Leontiadis, G.; Howden, C.W.; Moss, S.F. ACG clinical guideline: Treatment of Helicobacter pylori infection. Am. J. Gastroenterol. 2017, 112, 212–239. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, Z.; Hong, J.; Liu, W.; Lu, H.; Du, Y.; Wang, W.; Xu, J.; Wang, X.; Huo, L.; et al. Furazolidone-containing triple and quadruple eradication therapy for initial treatment for Helicobacter pylori infection: A multicenter randomized controlled trial in China. Helicobacter 2018, 23, e12496. [Google Scholar] [CrossRef]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.Y.; Crowe, S.E.; Valasek, M.A. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 2015, 43, 514–533. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017. Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 14 February 2019).
- Hu, Y.; Zhu, Y.; Lu, N.-H. Primary Antibiotic Resistance of Helicobacter pylori in China. Dig. Dis. Sci. 2017, 51, 70–1154. [Google Scholar] [CrossRef]
- Zobel, A.M.; Brown, S.A. Localization of daphnetin and umbelliferone in different tissues of Daphne mezereum shoots. Can. J. Bot. 1989, 67, 1456–1459. [Google Scholar] [CrossRef]
- Yang, Y.-Z.; Ranz, A.; Pan, H.-Z.; Zhang, Z.-N.; Lin, X.-B.; Meshnick, S.R. Daphnetin: A novel antimalarial agent with in vitro and in vivo activity. Am. J. Trop. Med. Hyg. 1992, 46, 15–20. [Google Scholar] [CrossRef]
- Fukuda, H.; Nakamura, S.; Chisaki, Y.; Takada, T.; Toda, Y.; Murata, H.; Itoh, K.; Yano, Y.; Takata, K.; Ashihara, E. Daphnetin inhibits invasion and migration of LM8 murine osteosarcoma cells by decreasing RhoA and Cdc42 expression. Biochem. Biophys. Res. Commun. 2016, 471, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhou, T.; Wang, J.; Sang, X.; Lan, L.; Luo, L.; Yin, Z. Daphnetin reduces endotoxin lethality in mice and decreases LPS-induced inflammation in Raw264.7 cells via suppressing JAK/STATs activation and ROS production. Inflamm. Res. 2017, 66, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Finn, G.J.; Creaven, B.S.; Egan, D.A. Daphnetin induced differentiation of human renal carcinoma cells and its mediation by p38 mitogen-activated protein kinase. Biochem. Pharmacol. 2004, 67, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Nanzhen, K.; Jieying, W.; Wenwei, Z.; Xiaoping, Z.; Yingyuan, F. Toxicological studies of daphnetin. Pharmacogn. Mag. 2018, 58, 561–566. [Google Scholar]
- Finn, G.J.; Kenealy, E.; Creaven, B.S.; Egan, D.A. In vitro cytotoxic potential and mechanism of action of selected coumarins, using human renal cell lines. Cancer Lett. 2002, 183, 61–68. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Camacho, D.M.; Kohanski, M.A.; Callura, J.M.; Collins, J.J. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 2012, 46, 561–572. [Google Scholar] [CrossRef]
- Duck, W.M.; Sobel, J.; Pruckler, J.M.; Song, Q.; Swerdlow, D.; Friedman, C.; Sulka, A.; Swaminathan, B.; Taylor, T.; Hoekstra, M.; et al. Antimicrobial resistance incidence and risk factors among Helicobacter pylori–infected persons, United States. Emerg. Infect. Dis. 2004, 10, 1088–1094. [Google Scholar] [CrossRef]
- De Francesco, V.; Giorgio, F.; Hassan, C.; Manes, G.; Vannella, L.; Panella, C.; Ierardi, E.; Zullo, A. Worldwide H. pylori antibiotic resistance: A systematic review. J. Gastrointestin. Liver Dis. 2010, 19, 409–414. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 5.0, 2015. 2015. Available online: http://www.eucast.org (accessed on 14 February 2019).
- Su, P.; Li, Y.; Li, H.; Zhang, J.; Lin, L.; Wang, Q.; Guo, F.; Ji, Z.; Mao, J.; Tang, W.; et al. Antibiotic resistance of Helicobacter Pylori isolated in the southeast coastal region of China. Helicobacter 2013, 18, 274–279. [Google Scholar] [CrossRef]
- De Francesco, V.; Zullo, A.; Fiorini, G.; Saracino, I.M.; Pavoni, M.; Vaira, D. Role of MIC levels of resistance to clarithromycin and metronidazole in Helicobacter pylori eradication. J. Antimicrob. Chemother. 2018. Available online: https://academic.oup.com/jac/advance-article-abstract/doi/10.1093/jac/dky469/5210026 (accessed on 26 November 2018). [CrossRef]
- Bai, P.; Zhou, L.Y.; Xiao, X.M.; Luo, Y.; Ding, Y. Susceptibility of Helicobacter pylorito antibiotics in Chinese patients. J. Dig. Dis. 2015, 16, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ding, W.; Xu, Y.; Wu, D.; Li, S.; Chen, J.; Guo, B. New insights into the antibacterial activity of Hydroxycoumarins against Ralstonia solanacearum. Molecules 2016, 21, 468. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.-U.; Khan, R.; Bhat, K.A.; Raja, A.F.; Shawl, A.S.; Alam, M.S. Isolation, characterisation and antibacterial activity studies of coumarins from Rhododendron lepidotum Wall. ex G. Don, Ericaceae. Rev. Bras. Farm. 2010, 20, 886–890. [Google Scholar] [CrossRef] [Green Version]
- Häcker, G. The morphology of apoptosis. Cell Tissue Res. 2000, 301, 5–17. [Google Scholar] [CrossRef]
- Allocati, N.; Masulli, M.; Di Ilio, C.; De Laurenzi, V. Die for the community: An overview of programmed cell death in bacteria. Cell Death Dis. 2015, 6, e1609. [Google Scholar] [CrossRef]
- Maraldi, N.M.; Cellini, L.; Robuffo, I.; Donelli, G. Searching the point of no return in Helicobacter pylori life: Necrosis and/or programmed death? J. Appl. Microbiol. 2001, 90, 727–732. [Google Scholar]
- Kusters, J.G.; Gerrits, M.M.; A Van Strijp, J.; Vandenbroucke-Grauls, C.M. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 1997, 65, 3672–3679. [Google Scholar] [PubMed]
- Shu, K.; Kuang, N.; Zhang, Z.; Hu, Z.; Zhang, Y.; Fu, Y.; Min, W. Therapeutic effect of daphnetin on the autoimmune arthritis through demethylation of proapoptotic genes in synovial cells. J. Transl. Med. 2014, 12, 287. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Bayles, K.W. Death’s toolbox: Examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 2003, 50, 729–738. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Wierzbowski, J.; Cottarel, G.; Collins, J.J. Mistranslation of Membrane Proteins and Two-Component System Activation Trigger Antibiotic-Mediated Cell Death. Cell 2008, 135, 679–690. [Google Scholar] [CrossRef] [Green Version]
- Erental, A.; Kalderon, Z.; Saada, A.; Smith, Y.; Engelberg-Kulka, H. Apoptosis-like death, an extreme SOS response in Escherichia coli. MBio 2014, 5, e01426-14. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Karasaki, M.; Ochiai, T.; Suzuki-Karasaki, Y. Crosstalk between mitochondrial ROS and depolarization in the potentiation of TRAIL-induced apoptosis in human tumor cells. Int. J. Oncol. 2013, 44, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Amundsen, S.K.; Fero, J.; Hansen, L.M.; Cromie, G.A.; Solnick, J.V.; Smith, G.R.; Salama, N.R. Helicobacter pyloriAddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 2008, 69, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Hage, N.; Howard, T. Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA. Sci Adv. 2015, 1, e1500315. [Google Scholar] [CrossRef] [PubMed]
- Strugatsky, D.; McNulty, R.; Munson, K.; Chen, C.-K.; Soltis, S.M.; Sachs, G.; Luecke, H. Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature 2012, 493, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Whitmire, J.M.; Merrell, D.S. Successful Culture Techniques for Helicobacter Species: General Culture Techniques for Helicobacter pylori. In Helicobacter Species; Humana Press: Totowa, NJ, USA, 2012; pp. 37–40. [Google Scholar]
- Cui, J.; Xing, L.; Li, Z.; Wu, S.; Wang, J.; Liu, J.; Wang, J.; Yan, X.; Zhang, X. Ochratoxin A induces G2 phase arrest in human gastric epithelium GES-1 cells in vitro. Toxicol. Lett. 2010, 193, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Best, L.M.; Haldane, D.J.M.; Keelan, M.; Taylor, D.E.; Thomson, A.B.R.; Loo, V.; Fallone, C.A.; Lyn, P.; Smaill, F.M.; Hunt, R.; et al. Multilaboratory Comparison of Proficiencies in Susceptibility Testing of Helicobacter pylori and correlation between agar dilution and E test methods. Antimicrob. Agents Chemother. 2003, 47, 3138–3144. [Google Scholar] [CrossRef] [PubMed]
- Obonyo, M.; Zhang, L.; Thamphiwatana, S.; Pornpattananangkul, D.; Fu, V.; Zhang, L. Antibacterial Activities of Liposomal Linolenic Acids against Antibiotic-Resistant Helicobacter pylori. Mol. Pharm. 2012, 9, 2677–2685. [Google Scholar] [CrossRef]
- Figura, N.; Marcolongo, R.; Cavallo, G.; Santucci, A.; Collodel, G.; Spreafico, A.; Moretti, E. Polysorbate 80 and Helicobacter pylori: A microbiological and ultrastructural study. BMC Microbiol. 2012, 12, 217. [Google Scholar] [CrossRef]
- Soo-Hwan, K.; Lee, H.S. Antibacterial Activity of Silver-nanoparticles Against Staphylococcus aureus and Escherichia coli. Korean J. Microbiol. Biotechnol. 2011, 39, 77–85. [Google Scholar]
- Dwyer, D.J.; A Kohanski, M.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Wan, C.; Xie, Q.; Huang, R.; Tao, X.; Shah, N.P.; Wei, H. Changes in gastric microbiota induced by Helicobacter pylori infection and preventive effects of Lactobacillus plantarum ZDY 2013 against such infection. J. Dairy Sci. 2016, 99, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Carlsohn, E.; Nyström, J.; Karlsson, H.; Svennerholm, A.-M.; Nilsson, C.L. Characterization of the outer membrane protein profile from disease-related Helicobacter pylori isolates by subcellular fractionation and nano-LC FT-ICR MS analysis. J. Proteome Res. 2006, 5, 3197–3204. [Google Scholar] [CrossRef] [PubMed]
- Baik, S.-C.; Kim, K.-M.; Song, S.-M.; Kim, D.-S.; Jun, J.-S.; Lee, S.-G.; Song, J.-Y.; Park, J.-U.; Kang, H.-L.; Lee, W.-K.; et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J. Bacteriol. 2004, 186, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, Y.; Pai, P.-J.; Chen, D.; Lam, H. Label-free quantitative proteomics analysis of antibiotic response in staphylococcus aureus to oxacillin. J. Proteome Res. 2014, 13, 1223–1233. [Google Scholar] [CrossRef] [PubMed]
- Domon, B.; Aebersold, R. Mass Spectrometry and Protein Analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Salas-Jara, M.J.; Sanhueza, E.A.; Retamal-Díaz, A.; González, C.; Urrutia, H.; García, A. Probiotic Lactobacillus fermentum UCO-979C biofilm formation on AGS and Caco-2 cells and Helicobacter pylori inhibition. Biofouling 2016, 32, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Niehues, M.; Euler, M.; Georgi, G.; Mank, M.; Stahl, B.; Hensel, A. Peptides from Pisum sativum L. enzymatic protein digest with anti-adhesive activity against Helicobacter pylori: Structure-activity and inhibitory activity against BabA, SabA, HpaA and a fibronectin-binding adhesin. Mol. Nutr. Food Res. 2010, 54, 1851–1861. [Google Scholar] [CrossRef]
- Li, N.; Han, L.; Chen, J.; Lin, X.; Chen, H.; She, F. Proliferative and apoptotic effects of gastric epithelial cells induced by coccoid Helicobacter pylori. J. Basic Microbiol. 2012, 53, 147–155. [Google Scholar] [CrossRef]






| Antibiotics | MICs (μg/mL) | Percent of Resistance (%) b | |
|---|---|---|---|
| 20 Clinical Isolates | ATCC43504 a | ||
| Daphnetin | 25–100 | 25 | NA c |
| Clarithromycin | 0.016–4 | 0.016 | 25% |
| Metronidazole | 4–256 | 128 | 85% |
| Groups | Mean of the Positive Fluorescence ± SD (%) | Protein Leakage (μg/mL) | ||
|---|---|---|---|---|
| PS Translocation | Membrane Permeability | Membrane Depolarization | ||
| Control | 5.93 ± 1.25 | 7.78 ± 0.62 | 9.26 ± 1.34 | 0.56 ± 0.01 |
| Daphnetin (12.5 μg/mL) | 56.99 ± 5.78 * | 5.06 ± 3.40 | 8.87 ± 2.71 | 0.60 ± 0.03 |
| Groups | Viability (mean ± SD/%) a | |
|---|---|---|
| DMEM | DMEM + 10% FBS b | |
| Control | 100.00 ± 7.79 | 100.00 ± 2.30 |
| Daphnetin 12.5 μg/mL | 84.43 ± 5.95 | 81.14 ± 11.52 |
| Primers | Sequence |
|---|---|
| ureI | Forward: CCCCTGTAGAAGGTGCTGAA Reverse: GCCGCATACAAGTAGGTGAAAC |
| babA | Forward: AAGCCTATCAAATCCTCCAAACG Reverse: TGGCGAGCAGTTATTATTCCCT |
| recA | Forward: CTAAGAGGTTGGGCGTGGA Reverse: CAATCCCTCCGCTTCTGGT |
| 16s rRNA | Forward: GTGCCAGCMGCCGCGGTAA Reverse: GACTACHVGGGTATCTAATCC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Pang, J.; Hu, X.; Nie, T.; Lu, X.; Li, X.; Wang, X.; Lu, Y.; Yang, X.; Jiang, J.; et al. Daphnetin: A Novel Anti-Helicobacter pylori Agent. Int. J. Mol. Sci. 2019, 20, 850. https://doi.org/10.3390/ijms20040850
Wang G, Pang J, Hu X, Nie T, Lu X, Li X, Wang X, Lu Y, Yang X, Jiang J, et al. Daphnetin: A Novel Anti-Helicobacter pylori Agent. International Journal of Molecular Sciences. 2019; 20(4):850. https://doi.org/10.3390/ijms20040850
Chicago/Turabian StyleWang, Genzhu, Jing Pang, Xinxin Hu, Tongying Nie, Xi Lu, Xue Li, Xiukun Wang, Yun Lu, Xinyi Yang, Jiandong Jiang, and et al. 2019. "Daphnetin: A Novel Anti-Helicobacter pylori Agent" International Journal of Molecular Sciences 20, no. 4: 850. https://doi.org/10.3390/ijms20040850
APA StyleWang, G., Pang, J., Hu, X., Nie, T., Lu, X., Li, X., Wang, X., Lu, Y., Yang, X., Jiang, J., Li, C., Xiong, Y. Q., & You, X. (2019). Daphnetin: A Novel Anti-Helicobacter pylori Agent. International Journal of Molecular Sciences, 20(4), 850. https://doi.org/10.3390/ijms20040850