Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors
Abstract
:1. Introduction
2. “Ecnomotopic” csGPCRs and Their Modulation by Small Molecules
2.1. Ecnomotopic ORs
2.2. Ecnomotopic Taste Receptors
2.3. Ecnomotopic Odorant and Taste GPCRs: A Functional Cross-Talk?
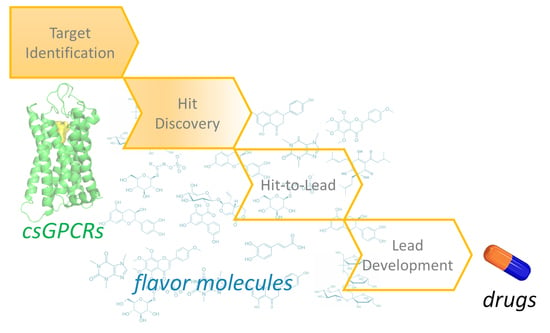
3. csGPCR Drug Discovery
3.1. csGPCR Hit Discovery
3.2. Drug-Likeness of Flavor Molecules
3.3. Towards the Discovery of csGPCR Lead Molecules
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GPCR | G protein-coupled receptor |
| csGPCR | chemosensory GPCR |
| TM | transmembrane |
| OR | odorant receptor |
| TAAR | trace amine-associated receptor |
| TAS2R | bitter taste receptor |
| TAS1R2/TAS1R3 | sweet taste receptor |
| TAS1R1/TAS1R3 | umami taste receptor |
| C4HSL | N-butanoyl-L-homoserine lactone |
| C12HSL | N-lauroyl-L-homoserine lactone |
| ASM | airway smooth muscle |
| GLP-1 | glucagon-like peptide 1 |
| MW | Molecular Weight |
| AlogP | octanol/water partition coefficient |
| HBD | Hydrogen Bond Donor |
| HBA | Hydrogen Bond Acceptor |
| RO5 | RULE OF 5 |
| CAAD | Computer-Aided Drug Design |
| QSAR | Quantitative Structure-Activity Relationship |
References
- Sriram, K.; Insel, P.A. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 2018, 93, 251–258. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829. [Google Scholar] [CrossRef]
- Davies, M.N.; Secker, A.; Halling-Brown, M.; Moss, D.S.; Freitas, A.A.; Timmis, J.; Clark, E.; Flower, D.R. GPCRTree: Online hierarchical classification of GPCR function. BMC Res. Notes 2008, 1, 67. [Google Scholar] [CrossRef]
- Pandy-Szekeres, G.; Munk, C.; Tsonkov, T.M.; Mordalski, S.; Harpsoe, K.; Hauser, A.S.; Bojarski, A.J.; Gloriam, D.E. GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res 2018, 46, D440–D446. [Google Scholar] [CrossRef]
- Harding, S.D.; Sharman, J.L.; Faccenda, E.; Southan, C.; Pawson, A.J.; Ireland, S.; Gray, A.J.G.; Bruce, L.; Alexander, S.P.H.; Anderton, S.; et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018, 46, D1091–D1106. [Google Scholar] [CrossRef]
- Cheng, A.C.; Coleman, R.G.; Smyth, K.T.; Cao, Q.; Soulard, P.; Caffrey, D.R.; Salzberg, A.C.; Huang, E.S. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 2007, 25, 71–75. [Google Scholar] [CrossRef]
- Jacobson, K.A. New paradigms in GPCR drug discovery. Biochem. Pharmacol. 2015, 98, 541–555. [Google Scholar] [CrossRef] [Green Version]
- Nieto Gutierrez, A.; McDonald, P.H. GPCRs: Emerging anti-cancer drug targets. Cell Signal. 2018, 41, 65–74. [Google Scholar] [CrossRef]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2016, 16, 19. [Google Scholar] [CrossRef]
- Ferrero, D.M.; Wacker, D.; Roque, M.A.; Baldwin, M.W.; Stevens, R.C.; Liberles, S.D. Agonists for 13 trace amine-associated receptors provide insight into the molecular basis of odor selectivity. ACS Chem. Biol. 2012, 7, 1184–1189. [Google Scholar] [CrossRef]
- Liberles, S.D.; Buck, L.B. A second class of chemosensory receptors in the olfactory epithelium. Nature 2006, 442, 645–650. [Google Scholar] [CrossRef]
- Gainetdinov, R.R.; Hoener, M.C.; Berry, M.D. Trace Amines and Their Receptors. Pharmacol. Rev. 2018, 70, 549–620. [Google Scholar] [CrossRef]
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.; Zuker, C.S. A novel family of mammalian taste receptors. Cell 2000, 100, 693–702. [Google Scholar] [CrossRef]
- Chandrashekar, J.; Mueller, K.L.; Hoon, M.A.; Adler, E.; Feng, L.X.; Guo, W.; Zuker, C.S.; Ryba, N.J.P. T2Rs function as bitter taste receptors. Cell 2000, 100, 703–711. [Google Scholar] [CrossRef]
- Matsunami, H.; Montmayeur, J.P.; Buck, L.B. A family of candidate taste receptors in human and mouse. Nature 2000, 404, 601–604. [Google Scholar] [CrossRef]
- Di Pizio, A.; Niv, M.Y. Computational Studies of Smell and Taste Receptors. Israel J. Chem. 2014, 54, 1205–1218. [Google Scholar] [CrossRef]
- Xu, H.; Staszewski, L.; Tang, H.; Adler, E.; Zoller, M.; Li, X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 14258–14263. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yao, Y.; Kuang, D.; Hampson, D.R. Activation of family C G-protein-coupled receptors by the tripeptide glutathione. J. Biol. Chem. 2006, 281, 8864–8870. [Google Scholar] [CrossRef]
- Li, X. T1R receptors mediate mammalian sweet and umami taste. Am. J. Clin. Nutr. 2009, 90, 733S–737S. [Google Scholar] [CrossRef]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef]
- Galindo, M.M.; Voigt, N.; Stein, J.; van Lengerich, J.; Raguse, J.D.; Hofmann, T.; Meyerhof, W.; Behrens, M. G Protein-Coupled Receptors in Human Fat Taste Perception. Chem. Senses 2012, 37, 123–139. [Google Scholar] [CrossRef]
- Offermanns, S. Free Fatty Acid (FFA) and Hydroxy Carboxylic Acid (HCA) Receptors. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 407–434. [Google Scholar] [CrossRef]
- Voigt, N.; Stein, J.; Galindo, M.M.; Dunkel, A.; Raguse, J.D.; Meyerhof, W.; Hofmann, T.; Behrens, M. The role of lipolysis in human orosensory fat perception. J. Lipid Res. 2014, 55, 870–882. [Google Scholar] [CrossRef] [Green Version]
- Ohsu, T.; Amino, Y.; Nagasaki, H.; Yamanaka, T.; Takeshita, S.; Hatanaka, T.; Maruyama, Y.; Miyamura, N.; Eto, Y. Involvement of the calcium-sensing receptor in human taste perception. J. Biol. Chem. 2010, 285, 1016–1022. [Google Scholar] [CrossRef]
- Dunkel, A.; Koster, J.; Hofmann, T. Molecular and sensory characterization of gamma-glutamyl peptides as key contributors to the kokumi taste of edible beans (Phaseolus vulgaris L.). J. Agric. Food Chem. 2007, 55, 6712–6719. [Google Scholar] [CrossRef]
- Krautwurst, D.; Kotthoff, M. A hit map-based statistical method to predict best ligands for orphan olfactory receptors: Natural key odorants versus "lock picks". Methods Mol. Biol. 2013, 1003, 85–97. [Google Scholar] [CrossRef]
- Dunkel, A.; Steinhaus, M.; Kotthoff, M.; Nowak, B.; Krautwurst, D.; Schieberle, P.; Hofmann, T. Nature’s chemical signatures in human olfaction: A foodborne perspective for future biotechnology. Angew. Chem. Int. Ed. Engl. 2014, 53, 7124–7143. [Google Scholar] [CrossRef]
- Foster, S.R.; Roura, E.; Thomas, W.G. Extrasensory perception: Odorant and taste receptors beyond the nose and mouth. Pharmacol. Ther. 2014, 142, 41–61. [Google Scholar] [CrossRef]
- Massberg, D.; Hatt, H. Human Olfactory Receptors: Novel Cellular Functions Outside of the Nose. Physiol. Rev. 2018, 98, 1739–1763. [Google Scholar] [CrossRef]
- Behrens, M.; Meyerhof, W. Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 2011, 105, 4–13. [Google Scholar] [CrossRef]
- Kang, N.; Koo, J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012, 45, 612–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2018. [Google Scholar] [CrossRef] [PubMed]
- Krautwurst, D. Preface: The chemical senses taste and smell in medicinal chemistry. In Taste and Smell; Krautwurst, D., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. V–XII. [Google Scholar]
- Roth, B.L.; Kroeze, W.K. Integrated Approaches for Genome-wide Interrogation of the Druggable Non-olfactory G Protein-coupled Receptor Superfamily. J. Biol. Chem. 2015, 290, 19471–19477. [Google Scholar] [CrossRef]
- Tautermann, C.S. GPCR structures in drug design, emerging opportunities with new structures. Bioorg. Med. Chem. Lett. 2014, 24, 4073–4079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Cruz, O.; Blekhman, R.; Zhang, X.; Nicolae, D.; Firestein, S.; Gilad, Y. A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes. Mol. Biol. Evol. 2009, 26, 491–494. [Google Scholar] [CrossRef]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef] [PubMed]
- Flegel, C.; Vogel, F.; Hofreuter, A.; Schreiner, B.S.; Osthold, S.; Veitinger, S.; Becker, C.; Brockmeyer, N.H.; Muschol, M.; Wennemuth, G.; et al. Characterization of the Olfactory Receptors Expressed in Human Spermatozoa. Front. Mol. Biosci. 2015, 2, 73. [Google Scholar] [CrossRef]
- Garcia-Esparcia, P.; Schluter, A.; Carmona, M.; Moreno, J.; Ansoleaga, B.; Torrejon-Escribano, B.; Gustincich, S.; Pujol, A.; Ferrer, I. Functional genomics reveals dysregulation of cortical olfactory receptors in Parkinson disease: Novel putative chemoreceptors in the human brain. J. Neuropathol. Exp. Neurol. 2013, 72, 524–539. [Google Scholar] [CrossRef]
- Aisenberg, W.H.; Huang, J.; Zhu, W.; Rajkumar, P.; Cruz, R.; Santhanam, L.; Natarajan, N.; Yong, H.M.; De Santiago, B.; Oh, J.J.; et al. Defining an olfactory receptor function in airway smooth muscle cells. Sci. Rep. 2016, 6, 38231. [Google Scholar] [CrossRef] [Green Version]
- Spehr, M.; Gisselmann, G.; Poplawski, A.; Riffell, J.A.; Wetzel, C.H.; Zimmer, R.K.; Hatt, H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 2003, 299, 2054–2058. [Google Scholar] [CrossRef]
- Braun, T.; Voland, P.; Kunz, L.; Prinz, C.; Gratzl, M. Enterochromaffin cells of the human gut: Sensors for spices and odorants. Gastroenterology 2007, 132, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Grison, A.; Zucchelli, S.; Urzi, A.; Zamparo, I.; Lazarevic, D.; Pascarella, G.; Roncaglia, P.; Giorgetti, A.; Garcia-Esparcia, P.; Vlachouli, C.; et al. Mesencephalic dopaminergic neurons express a repertoire of olfactory receptors and respond to odorant-like molecules. BMC Genomics 2014, 15, 729. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.; Veitinger, S.; Peek, I.; Busse, D.; Eckardt, J.; Vladimirova, D.; Jovancevic, N.; Wojcik, S.; Gisselmann, G.; Altmuller, J.; et al. Two olfactory receptors-OR2A4/7 and OR51B5-differentially affect epidermal proliferation and differentiation. Exp. Dermatol. 2017, 26, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Marcinek, P.; Geithe, C.; Krautwurst, D. Chemosensory G Protein-Coupled Receptors (GPCR) in Blood Leukocytes. In Taste and Smell; Krautwurst, D., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 151–173. [Google Scholar]
- Behrens, M.; Meyerhof, W. Oral and extraoral bitter taste receptors. Results Probl. Cell Differ. 2010, 52, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.R.; Blank, K.; See Hoe, L.E.; Behrens, M.; Meyerhof, W.; Peart, J.N.; Thomas, W.G. Bitter taste receptor agonists elicit G-protein-dependent negative inotropy in the murine heart. FASEB J. 2014, 28, 4497–4508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sternini, C.; Anselmi, L.; Rozengurt, E. Enteroendocrine cells: A site of “taste“ in gastrointestinal chemosensing. Curr. Opin. Endocrinol. Diabetes Obes. 2008, 15, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Latorre, R.; Huynh, J.; Mazzoni, M.; Gupta, A.; Bonora, E.; Clavenzani, P.; Chang, L.; Mayer, E.A.; De Giorgio, R.; Sternini, C. Expression of the Bitter Taste Receptor, T2R38, in Enteroendocrine Cells of the Colonic Mucosa of Overweight/Obese vs. Lean Subjects. PLoS ONE 2016, 11, e0147468. [Google Scholar] [CrossRef]
- Carey, R.M.; Adappa, N.D.; Palmer, J.N.; Lee, R.J.; Cohen, N.A. Taste Receptors: Regulators of Sinonasal Innate Immunity. Laryngoscope Investig. Otolaryngol. 2016, 1, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Malki, A.; Fiedler, J.; Fricke, K.; Ballweg, I.; Pfaffl, M.W.; Krautwurst, D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J. Leukoc. Biol. 2015, 97, 533–545. [Google Scholar] [CrossRef] [Green Version]
- Harrington, E.O.; Vang, A.; Braza, J.; Shil, A.; Chichger, H. Activation of the sweet taste receptor, T1R3, by the artificial sweetener sucralose regulates the pulmonary endothelium. Am. J. Physiol.-Lung C 2018, 314, L165–L176. [Google Scholar] [CrossRef]
- Calvo, S.S.; Egan, J.M. The endocrinology of taste receptors. Nat. Rev. Endocrinol. 2015, 11, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Shaw, L.; Mansfield, C.; Colquitt, L.; Lin, C.L.; Ferreira, J.; Emmetsberger, J.; Reed, D.R. Personalized expression of bitter ‘taste’ receptors in human skin. PLoS ONE 2018, 13, e0205322. [Google Scholar] [CrossRef]
- Behrens, M.; Meyerhof, W. A role for taste receptors in (neuro)endocrinology? J. Neuroendocrinol. 2019, e12691. [Google Scholar] [CrossRef]
- Feldmesser, E.; Olender, T.; Khen, M.; Yanai, I.; Ophir, R.; Lancet, D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics 2006, 7, 121. [Google Scholar] [CrossRef]
- Medical Dictionary. Available online: https://medical-dictionary.thefreedictionary.com/nomotopic (accessed on 24 January 2019).
- Jaggupilli, A.; Singh, N.; Upadhyaya, J.; Sikarwar, A.S.; Arakawa, M.; Dakshinamurti, S.; Bhullar, R.P.; Duan, K.; Chelikani, P. Analysis of the expression of human bitter taste receptors in extraoral tissues. Mol. Cell. Biochem. 2017, 426, 137–147. [Google Scholar] [CrossRef]
- Olender, T.; Keydar, I.; Pinto, J.M.; Tatarskyy, P.; Alkelai, A.; Chien, M.S.; Fishilevich, S.; Restrepo, D.; Matsunami, H.; Gilad, Y.; et al. The human olfactory transcriptome. BMC Genomics 2016, 17, 619. [Google Scholar] [CrossRef]
- Douglas, J.E.; Lin, C.; Mansfield, C.J.; Arayata, C.J.; Cowart, B.J.; Spielman, A.I.; Adappa, N.D.; Palmer, J.N.; Cohen, N.A.; Reed, D.R. Tissue-Dependent Expression of Bitter Receptor TAS2R38 mRNA. Chem. Senses 2019, 44, 33–40. [Google Scholar] [CrossRef]
- Lu, P.; Zhang, C.H.; Lifshitz, L.M.; ZhuGe, R. Extraoral bitter taste receptors in health and disease. J. Gen. Physiol. 2017, 149, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Shaik, F.A.; Singh, N.; Arakawa, M.; Duan, K.; Bhullar, R.P.; Chelikani, P. Bitter taste receptors: Extraoral roles in pathophysiology. Int. J. Biochem. Cell Biol. 2016, 77, 197–204. [Google Scholar] [CrossRef]
- Ferrer, I.; Garcia-Esparcia, P.; Carmona, M.; Carro, E.; Aronica, E.; Kovacs, G.G.; Grison, A.; Gustincich, S. Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front. Aging Neurosci. 2016, 8, 163. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, H.; Fu, N.; Chen, L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J. Cell. Physiol. 2018, 233, 2104–2115. [Google Scholar] [CrossRef]
- Clark, A.A.; Dotson, C.D.; Elson, A.E.; Voigt, A.; Boehm, U.; Meyerhof, W.; Steinle, N.I.; Munger, S.D. TAS2R bitter taste receptors regulate thyroid function. FASEB J. 2015, 29, 164–172. [Google Scholar] [CrossRef]
- Christian, S.L.; Berry, M.D. Trace Amine-Associated Receptors as Novel Therapeutic Targets for Immunomodulatory Disorders. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef]
- Dalesio, N.M.; Ortiz, S.F.B.; Pluznick, J.L.; Berkowitz, D.E. Olfactory, Taste, and Photo Sensory Receptors in Non-sensory Organs: It Just Makes Sense. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Cheret, J.; Bertolini, M.; Ponce, L.; Lehmann, J.; Tsai, T.; Alam, M.; Hatt, H.; Paus, R. Olfactory receptor OR2AT4 regulates human hair growth. Nat. Commun. 2018, 9, 3624. [Google Scholar] [CrossRef]
- Manteniotis, S.; Wojcik, S.; Brauhoff, P.; Mollmann, M.; Petersen, L.; Gothert, J.R.; Schmiegel, W.; Duhrsen, U.; Gisselmann, G.; Hatt, H. Functional characterization of the ectopically expressed olfactory receptor 2AT4 in human myelogenous leukemia. Cell Death Discov. 2016, 2, 15070. [Google Scholar] [CrossRef] [Green Version]
- Gelis, L.; Jovancevic, N.; Bechara, F.G.; Neuhaus, E.M.; Hatt, H. Functional expression of olfactory receptors in human primary melanoma and melanoma metastasis. Exp. Dermatol. 2017, 26, 569–576. [Google Scholar] [CrossRef]
- Cui, T.; Tsolakis, A.V.; Li, S.C.; Cunningham, J.L.; Lind, T.; Oberg, K.; Giandomenico, V. Olfactory receptor 51E1 protein as a potential novel tissue biomarker for small intestine neuroendocrine carcinomas. Eur. J. Endocrinol. 2013, 168, 253–261. [Google Scholar] [CrossRef]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory Receptor Family 7 Subfamily C Member 1 Is a Novel Marker of Colon Cancer-Initiating Cells and Is a Potent Target of Immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef] [Green Version]
- Weber, L.; Massberg, D.; Becker, C.; Altmuller, J.; Ubrig, B.; Bonatz, G.; Wolk, G.; Philippou, S.; Tannapfel, A.; Hatt, H.; et al. Olfactory Receptors as Biomarkers in Human Breast Carcinoma Tissues. Front. Oncol. 2018, 8, 33. [Google Scholar] [CrossRef]
- Ranzani, M.; Iyer, V.; Ibarra-Soria, X.; Del Castillo Velasco-Herrera, M.; Garnett, M.; Logan, D.; Adams, D.J. Revisiting olfactory receptors as putative drivers of cancer. Wellcome Open Res. 2017, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, M.; Siwko, S.; Liu, M. Prostate-Specific G-Protein Coupled Receptor, an Emerging Biomarker Regulating Inflammation and Prostate Cancer Invasion. Curr. Mol. Med. 2016, 16, 526–532. [Google Scholar] [CrossRef]
- Cao, W.Q.; Li, F.Q.; Yao, J.G.; Yu, J.Z. Prostate specific G protein coupled receptor is associated with prostate cancer prognosis and affects cancer cell proliferation and invasion. BMC Cancer 2015, 15, 915. [Google Scholar] [CrossRef]
- Sanz, G.; Leray, I.; Dewaele, A.; Sobilo, J.; Lerondel, S.; Bouet, S.; Grebert, D.; Monnerie, R.; Pajot-Augy, E.; Mir, L.M. Promotion of Cancer Cell Invasiveness and Metastasis Emergence Caused by Olfactory Receptor Stimulation. PLoS ONE 2014, 9, e85110. [Google Scholar] [CrossRef]
- Neuhaus, E.M.; Zhang, W.Y.; Gelis, L.; Deng, Y.; Noldus, J.; Hatt, H. Activation of an Olfactory Receptor Inhibits Proliferation of Prostate Cancer Cells. J. Biol. Chem. 2009, 284, 16218–16225. [Google Scholar] [CrossRef]
- Abaffy, T.; Bain, J.R.; Muehlbauer, M.J.; Spasojevic, I.; Lodha, S.; Bruguera, E.; O’Neal, S.K.; Kim, S.Y.; Matsunami, H. A Testosterone Metabolite 19-Hydroxyandrostenedione Induces Neuroendocrine Trans-Differentiation of Prostate Cancer Cells via an Ectopic Olfactory Receptor. Front. Oncol. 2018, 8, 162. [Google Scholar] [CrossRef]
- Marcu, M.; Radu, E.; Sajin, M. Neuroendocrine transdifferentiation of prostate carcinoma cells and its prognostic significance. Rom. J. Morphol. Embryol. 2010, 51, 7–12. [Google Scholar]
- Massberg, D.; Jovancevic, N.; Offermann, A.; Simon, A.; Baniahmad, A.; Perner, S.; Pungsrinont, T.; Luko, K.; Philippou, S.; Ubrig, B.; et al. The activation of OR51E1 causes growth suppression of human prostate cancer cells. Oncotarget 2016, 7, 48231–48249. [Google Scholar] [CrossRef] [Green Version]
- Kalbe, B.; Schulz, V.M.; Schlimm, M.; Philippou, S.; Jovancevic, N.; Jansen, F.; Scholz, P.; Lubbert, H.; Jarocki, M.; Faissner, A.; et al. Helional-induced activation of human olfactory receptor 2J3 promotes apoptosis and inhibits proliferation in a non-small-cell lung cancer cell line. Eur. J. Cell Biol. 2017, 96, 34–46. [Google Scholar] [CrossRef]
- Weber, L.; Al-Refae, K.; Ebbert, J.; Jagers, P.; Altmuller, J.; Becker, C.; Hahn, S.; Gisselmann, G.; Hatt, H. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLoS ONE 2017, 12, e0172491. [Google Scholar] [CrossRef]
- Wang, Q.; Liszt, K.I.; Deloose, E.; Canovai, E.; Thijs, T.; Farre, R.; Ceulemans, L.J.; Lannoo, M.; Tack, J.; Depoortere, I. Obesity alters adrenergic and chemosensory signaling pathways that regulate ghrelin secretion in the human gut. FASEB J. 2019. [Google Scholar] [CrossRef]
- Bellono, N.W.; Bayrer, J.R.; Leitch, D.B.; Castro, J.; Zhang, C.; O‘Donnell, T.A.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell 2017, 170, 185–198.e116. [Google Scholar] [CrossRef]
- Jang, H.J.; Kokrashvili, Z.; Theodorakis, M.J.; Carlson, O.D.; Kim, B.J.; Zhou, J.; Kim, H.H.; Xu, X.; Chan, S.L.; Juhaszova, M.; et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc. Natl. Acad. Sci. USA 2007, 104, 15069–15074. [Google Scholar] [CrossRef] [Green Version]
- Steinert, R.E.; Gerspach, A.C.; Gutmann, H.; Asarian, L.; Drewe, J.; Beglinger, C. The functional involvement of gut-expressed sweet taste receptors in glucose-stimulated secretion of glucagon-like peptide-1 (GLP-1) and peptide YY (PYY). Clin. Nutr. 2011, 30, 524–532. [Google Scholar] [CrossRef]
- Ma, J.; Bellon, M.; Wishart, J.M.; Young, R.; Blackshaw, L.A.; Jones, K.L.; Horowitz, M.; Rayner, C.K. Effect of the artificial sweetener, sucralose, on gastric emptying and incretin hormone release in healthy subjects. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G735–G739. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Wideman, R.D.; Speck, M.; Asadi, A.; King, D.S.; Webber, T.D.; Haneda, M.; Kieffer, T.J. Incretin release from gut is acutely enhanced by sugar but not by sweeteners in vivo. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E473–G479. [Google Scholar] [CrossRef]
- Sylvetsky, A.C.; Brown, R.J.; Blau, J.E.; Walter, M.; Rother, K.I. Hormonal responses to non-nutritive sweeteners in water and diet soda. Nutr. Metab. (Lond.) 2016, 13, 71. [Google Scholar] [CrossRef]
- Saltiel, M.Y.; Kuhre, R.E.; Christiansen, C.B.; Eliasen, R.; Conde-Frieboes, K.W.; Rosenkilde, M.M.; Holst, J.J. Sweet Taste Receptor Activation in the Gut Is of Limited Importance for Glucose-Stimulated GLP-1 and GIP Secretion. Nutrients 2017, 9, 418. [Google Scholar] [CrossRef]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.K.; Vigues, S.; Ott, S.H.; Elson, A.E.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter taste receptors influence glucose homeostasis. PLoS ONE 2008, 3, e3974. [Google Scholar] [CrossRef]
- Kim, K.S.; Egan, J.M.; Jang, H.J. Denatonium induces secretion of glucagon-like peptide-1 through activation of bitter taste receptor pathways (vol. 57, pg 2117, 2014). Diabetologia 2014, 57, 2428. [Google Scholar] [CrossRef]
- Kok, B.P.; Galmozzi, A.; Littlejohn, N.K.; Albert, V.; Godio, C.; Kim, W.; Kim, S.M.; Bland, J.S.; Grayson, N.; Fang, M.; et al. Intestinal bitter taste receptor activation alters hormone secretion and imparts metabolic benefits. Mol. Metab. 2018, 16, 76–87. [Google Scholar] [CrossRef]
- Deloose, E.; Janssen, P.; Corsetti, M.; Biesiekierski, J.; Masuy, I.; Rotondo, A.; Van Oudenhove, L.; Depoortere, I.; Tack, J. Intragastric infusion of denatonium benzoate attenuates interdigestive gastric motility and hunger scores in healthy female volunteers. Am. J. Clin. Nutr. 2017, 105, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Janssen, S.; Laermans, J.; Verhulst, P.J.; Thijs, T.; Tack, J.; Depoortere, I. Bitter taste receptors and alpha-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying. Proc. Natl. Acad. Sci. USA 2011, 108, 2094–2099. [Google Scholar] [CrossRef]
- Meyer-Gerspach, A.C.; Biesiekierski, J.R.; Deloose, E.; Clevers, E.; Rotondo, A.; Rehfeld, J.F.; Depoortere, I.; Van Oudenhove, L.; Tack, J. Effects of caloric and noncaloric sweeteners on antroduodenal motility, gastrointestinal hormone secretion and appetite-related sensations in healthy subjects. Am. J. Clin. Nutr. 2018, 107, 707–716. [Google Scholar] [CrossRef]
- Tizzano, M.; Gulbransen, B.D.; Vandenbeuch, A.; Clapp, T.R.; Herman, J.P.; Sibhatu, H.M.; Churchill, M.E.A.; Silver, W.L.; Kinnamon, S.C.; Finger, T.E. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl. Acad. Sci. USA 2010, 107, 3210–3215. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.J.; Xiong, G.; Kofonow, J.M.; Chen, B.; Lysenko, A.; Jiang, P.; Abraham, V.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Investig. 2012, 122, 4145–4159. [Google Scholar] [CrossRef] [Green Version]
- Verbeurgt, C.; Veithen, A.; Carlot, S.; Tarabichi, M.; Dumont, J.E.; Hassid, S.; Chatelain, P. The human bitter taste receptor T2R38 is broadly tuned for bacterial compounds. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Hariri, B.M.; McMahon, D.B.; Chen, B.; Freund, J.R.; Mansfield, C.J.; Doghramji, L.J.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Reed, D.R.; et al. Flavones modulate respiratory epithelial innate immunity: Anti-inflammatory effects and activation of the T2R14 receptor. J. Biol. Chem. 2017, 292, 8484–8497. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.; Cohen, N. Taste receptors in innate immunity. Cell. Mol. Life Sci. 2015, 72, 217–236. [Google Scholar] [CrossRef]
- Lee, R.J.; Kofonow, J.M.; Rosen, P.L.; Siebert, A.P.; Chen, B.; Doghramji, L.; Xiong, G.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; et al. Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J. Clin. Investig. 2014, 124, 1393–1405. [Google Scholar] [CrossRef] [Green Version]
- Ekoff, M.; Choi, J.H.; James, A.; Dahlen, B.; Nilsson, G.; Dahlen, S.E. Bitter taste receptor (TAS2R) agonists inhibit IgE-dependent mast cell activation. J. Allergy Clin. Immunol. 2014, 134, 475–478. [Google Scholar] [CrossRef]
- Babusyte, A.; Kotthoff, M.; Fiedler, J.; Krautwurst, D. Biogenic amines activate blood leukocytes via trace amine-associated receptors TAAR1 and TAAR2. J. Leukoc. Biol. 2013, 93, 387–394. [Google Scholar] [CrossRef]
- D’Andrea, G.; Terrazzino, S.; Fortin, D.; Farruggio, A.; Rinaldi, L.; Leon, A. HPLC electrochemical detection of trace amines in human plasma and platelets and expression of mRNA transcripts of trace amine receptors in circulating leukocytes. Neurosci. Lett. 2003, 346, 89–92. [Google Scholar] [CrossRef]
- Geithe, C.; Andersen, G.; Malki, A.; Krautwurst, D. A Butter Aroma Recombinate Activates Human Class-I Odorant Receptors. J. Agric. Food Chem. 2015, 63, 9410–9420. [Google Scholar] [CrossRef]
- Belvisi, M.G.; Dale, N.; Birrell, M.A.; Canning, B.J. Bronchodilator activity of bitter tastants in human tissue. Nat. Med. 2011, 17, 776. [Google Scholar] [CrossRef]
- Deshpande, D.A.; Robinett, K.S.; Wang, W.C.; Sham, J.S.; An, S.S.; Liggett, S.B. Bronchodilator activity of bitter tastants in human tissue. Nat. Med. 2011, 17, 776–778. [Google Scholar] [CrossRef]
- Deshpande, D.A.; Wang, W.C.; McIlmoyle, E.L.; Robinett, K.S.; Schillinger, R.M.; An, S.S.; Sham, J.S.; Liggett, S.B. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat. Med. 2010, 16, 1299–1304. [Google Scholar] [CrossRef]
- An, S.S.; Liggett, S.B. Taste and smell GPCRs in the lung: Evidence for a previously unrecognized widespread chemosensory system. Cell Signal. 2018, 41, 82–88. [Google Scholar] [CrossRef]
- Toelstede, S.; Hofmann, T. Sensomics mapping and identification of the key bitter metabolites in Gouda cheese. J. Agric. Food Chem. 2008, 56, 2795–2804. [Google Scholar] [CrossRef]
- Intelmann, D.; Haseleu, G.; Dunkel, A.; Lagemann, A.; Stephan, A.; Hofmann, T. Comprehensive sensomics analysis of hop-derived bitter compounds during storage of beer. J. Agric. Food Chem. 2011, 59, 1939–1953. [Google Scholar] [CrossRef]
- Hillmann, H.; Mattes, J.; Brockhoff, A.; Dunkel, A.; Meyerhof, W.; Hofmann, T. Sensomics analysis of taste compounds in balsamic vinegar and discovery of 5-acetoxymethyl-2-furaldehyde as a novel sweet taste modulator. J. Agric. Food Chem. 2012, 60, 9974–9990. [Google Scholar] [CrossRef]
- Kiefl, J.; Pollner, G.; Schieberle, P. Sensomics analysis of key hazelnut odorants (Corylus avellana L. “Tonda Gentile“) using comprehensive two-dimensional gas chromatography in combination with time-of-flight mass spectrometry (GCxGC-TOF-MS). J. Agric. Food Chem. 2013, 61, 5226–5235. [Google Scholar] [CrossRef]
- Uselmann, V.; Schieberle, P. Decoding the combinatorial aroma code of a commercial Cognac by application of the sensomics concept and first insights into differences from a German brandy. J. Agric. Food Chem. 2015, 63, 1948–1956. [Google Scholar] [CrossRef]
- Meyer, S.; Dunkel, A.; Hofmann, T. Sensomics-Assisted Elucidation of the Tastant Code of Cooked Crustaceans and Taste Reconstruction Experiments. J. Agric. Food Chem. 2016, 64, 1164–1175. [Google Scholar] [CrossRef]
- Kranz, M.; Viton, F.; Smarrito-Menozzi, C.; Hofmann, T. Sensomics-Based Molecularization of the Taste of Pot-au-Feu, a Traditional Meat/Vegetable Broth. J. Agric. Food Chem. 2018, 66, 194–202. [Google Scholar] [CrossRef]
- Meyerhof, W.; Batram, C.; Kuhn, C.; Brockhoff, A.; Chudoba, E.; Bufe, B.; Appendino, G.; Behrens, M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 2010, 35, 157–170. [Google Scholar] [CrossRef]
- Noe, F.; Polster, J.; Geithe, C.; Kotthoff, M.; Schieberle, P.; Krautwurst, D. OR2M3: A Highly Specific and Narrowly Tuned Human Odorant Receptor for the Sensitive Detection of Onion Key Food Odorant 3-Mercapto-2-methylpentan-1-ol. Chem. Senses 2017, 42, 195–210. [Google Scholar] [CrossRef]
- Mainland, J.D.; Li, Y.R.; Zhou, T.; Liu, W.L.; Matsunami, H. Human olfactory receptor responses to odorants. Sci. Data 2015, 2, 150002. [Google Scholar] [CrossRef]
- Wallrabenstein, I.; Kuklan, J.; Weber, L.; Zborala, S.; Werner, M.; Altmuller, J.; Becker, C.; Schmidt, A.; Hatt, H.; Hummel, T.; et al. Human Trace Amine-Associated Receptor TAAR5 Can Be Activated by Trimethylamine. PLoS ONE 2013, 8, e54950. [Google Scholar] [CrossRef]
- Suess, B.; Brockhoff, A.; Degenhardt, A.; Billmayer, S.; Meyerhof, W.; Hofmann, T. Human Taste and Umami Receptor Responses to Chemosensorica Generated by Maillard-type N-2-Alkyl- and N-2-Arylthiomethylation of Guanosine 5-Monophosphates. J. Agric. Food Chem. 2014, 62, 11429–11440. [Google Scholar] [CrossRef]
- Bastiaan-Net, S.; van de Berg-Somhorst, B.P.M.; Ariens, R.M.C.; Paques, M.; Mes, J.J. A novel functional screening assay to monitor sweet taste receptor activation in vitro. Chem. Senses 2017, 42, E20–E21. [Google Scholar] [CrossRef]
- Geithe, C.; Noe, F.; Kreissl, J.; Krautwurst, D. The Broadly Tuned Odorant Receptor OR1A1 is Highly Selective for 3-Methyl-2,4-nonanedione, a Key Food Odorant in Aged Wines, Tea, and Other Foods. Chem. Senses 2017, 42, 181–193. [Google Scholar] [CrossRef]
- Thalmann, S.; Behrens, M.; Meyerhof, W. Major haplotypes of the human bitter taste receptor TAS2R41 encode functional receptors for chloramphenicol. Biochem. Biophys. Res. Commun. 2013, 435, 267–273. [Google Scholar] [CrossRef]
- Lossow, K.; Hubner, S.; Roudnitzky, N.; Slack, J.P.; Pollastro, F.; Behrens, M.; Meyerhof, W. Comprehensive Analysis of Mouse Bitter Taste Receptors Reveals Different Molecular Receptive Ranges for Orthologous Receptors in Mice and Humans. J. Biol. Chem. 2016, 291, 15358–15377. [Google Scholar] [CrossRef]
- Di Pizio, A.; Ben Shoshan-Galeczki, Y.; Hayes, J.E.; Niv, M.Y. Bitter and sweet tasting molecules: It’s complicated. Neurosci. Lett. 2018. [Google Scholar] [CrossRef]
- Behrens, M.; Meyerhof, W.; Hellfritsch, C.; Hofmann, T. Sweet and umami taste: Natural products, their chemosensory targets, and beyond. Angew. Chem. Int. Ed. Engl. 2011, 50, 2220–2242. [Google Scholar] [CrossRef]
- Winkel, C.; de Klerk, A.; Visser, J.; de Rijke, E.; Bakker, J.; Koenig, T.; Renes, H. New developments in umami (enhancing) molecules. Chem. Biodivers. 2008, 5, 1195–1203. [Google Scholar] [CrossRef]
- Suess, B.; Festring, D.; Hofmann, T. Umami compounds and taste enhancers. In Flavour Development, Analysis and Perception in Food and Beverages; Parker, K., Elmore, S., Methven, L., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; pp. 331–351. [Google Scholar]
- Dagan-Wiener, A.; Nissim, I.; Ben Abu, N.; Borgonovo, G.; Bassoli, A.; Niv, M.Y. Bitter or not? BitterPredict, a tool for predicting taste from chemical structure. Sci. Rep. 2017, 7, 12074. [Google Scholar] [CrossRef] [Green Version]
- Di Pizio, A.; Niv, M.Y. Promiscuity and selectivity of bitter molecules and their receptors. Bioorg. Med. Chem. 2015, 23, 4082–4091. [Google Scholar] [CrossRef]
- Fletcher, J.N.; Pan, L.; Kinghorn, A.D. Medicinal Chemistry of Plant Naturals as Agonists/Antagonists for Taste Receptors. In Taste and Smell; Krautwurst, D., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 35–71. [Google Scholar]
- Di Pizio, A.; Levit, A.; Slutzki, M.; Behrens, M.; Karaman, R.; Niv, M.Y. Comparing Class A GPCRs to bitter taste receptors: Structural motifs, ligand interactions and agonist-to-antagonist ratios. Methods Cell Biol. 2016, 132, 401–427. [Google Scholar] [CrossRef]
- Keller, T.H.; Pichota, A.; Yin, Z. A practical view of “druggability”. Curr. Opin. Chem. Biol. 2006, 10, 357–361. [Google Scholar] [CrossRef]
- Walters, W.P.; Murcko, A.; Murcko, M.A. Recognizing molecules with drug-like properties. Curr. Opin. Chem. Biol. 1999, 3, 384–387. [Google Scholar] [CrossRef]
- Oprea, T.I. Property distribution of drug-related chemical databases. J. Comput. Aided Mol. Des. 2000, 14, 251–264. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [Green Version]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. Properties of known drugs. 2. Side chains. J. Med. Chem. 1999, 42, 5095–5099. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ursu, O.; Rayan, A.; Goldblum, A.; Oprea, T.I. Understanding drug-likeness. Wires Comput. Mol. Sci. 2011, 1, 760–781. [Google Scholar] [CrossRef]
- Leeson, P.D.; Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 2007, 6, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Dagan-Wiener, A.; Di Pizio, A.; Nissim, I.; Bahia, M.S.; Dubovski, N.; Margulis, E.; Niv, M.Y. BitterDB: Taste ligands and receptors database in 2019. Nucleic Acids Res 2018, 47, D1179–D1185. [Google Scholar] [CrossRef] [PubMed]
- Wiener, A.; Shudler, M.; Levit, A.; Niv, M.Y. BitterDB: A database of bitter compounds. Nucleic Acids Res. 2012, 40, D413–D419. [Google Scholar] [CrossRef] [PubMed]
- natural and artificial sweetening agents. Nucleic Acids Res. 2011, 39, D377–D382. [CrossRef] [PubMed]
- Kim, N.C.; Kinghorn, A.D. Highly sweet compounds of plant origin. Arch. Pharm. Res. 2002, 25, 725–746. [Google Scholar] [CrossRef]
- Amino, Y.; Tahara, Y.; Yamada, K.; Nakazawa, M.; Tagami, U.; Tajima, T.; Kuroda, M. Design, synthesis, and taste evaluation of a high-intensity umami-imparting oxazole-based compound. Biosci. Biotechnol. Biochem. 2017, 81, 1690–1698. [Google Scholar] [CrossRef]
- Backes, M.; Paetz, S.; Vossing, T.; Ley, J.P. Synthesis and sensory studies of umami-active scaffolds. Chem. Biodivers. 2014, 11, 1782–1797. [Google Scholar] [CrossRef]
- Nissim, I.; Dagan-Wiener, A.; Niv, M.Y. The taste of toxicity: A quantitative analysis of bitter and toxic molecules. IUBMB Life 2017, 69, 938–946. [Google Scholar] [CrossRef]
- Cho, M.J.; Unklesbay, N.; Hsieh, F.H.; Clarke, A.D. Hydrophobicity of bitter peptides from soy protein hydrolysates. J. Agric. Food Chem. 2004, 52, 5895–5901. [Google Scholar] [CrossRef]
- Horne, J.; Lawless, H.T.; Speirs, W.; Sposato, D. Bitter taste of saccharin and acesulfame-K. Chem. Senses 2002, 27, 31–38. [Google Scholar] [CrossRef]
- Winnig, M.; Kuhn, C.; Frank, O.; Bufe, B.; Behrens, M.; Hofmann, T.; Meyerhof, W. Saccharin: Artificial Sweetener, Bitter Tastant, and Sweet Taste Inhibitor. ACS Symp. Ser. 2008, 979, 230–240. [Google Scholar] [CrossRef]
- Hellfritsch, C.; Brockhoff, A.; Stahler, F.; Meyerhof, W.; Hofmnann, T. Human Psychometric and Taste Receptor Responses to Steviol Glycosides. J. Agric. Food Chem. 2012, 60, 6782–6793. [Google Scholar] [CrossRef]
- Behrens, M.; Blank, K.; Meyerhof, W. Blends of Non-caloric Sweeteners Saccharin and Cyclamate Show Reduced Off-Taste due to TAS2R Bitter Receptor Inhibition. Cell Chem. Biol. 2017, 24, 1199–1204.e2. [Google Scholar] [CrossRef]
- Kuhn, C.; Bufe, B.; Winnig, M.; Hofmann, T.; Frank, O.; Behrens, M.; Lewtschenko, T.; Slack, J.P.; Ward, C.D.; Meyerhof, W. Bitter taste receptors for saccharin and acesulfame K. J. Neurosci. 2004, 24, 10260–10265. [Google Scholar] [CrossRef]
- Suess, B.; Brockhoff, A.; Meyerhof, W.; Hofmann, T. The Odorant (R)-Citronellal Attenuates Caffeine Bitterness by Inhibiting the Bitter Receptors TAS2R43 and TAS2R46. J. Agric. Food Chem. 2018, 66, 2301–2311. [Google Scholar] [CrossRef]
- Hopkins, A.L.; Groom, C.R.; Alex, A. Ligand efficiency: A useful metric for lead selection. Drug Discov. Today 2004, 9, 430–431. [Google Scholar] [CrossRef]
- Ryckmans, T.; Edwards, M.P.; Horne, V.A.; Correia, A.M.; Owen, D.R.; Thompson, L.R.; Tran, I.; Tutt, M.F.; Young, T. Rapid assessment of a novel series of selective CB(2) agonists using parallel synthesis protocols: A Lipophilic Efficiency (LipE) analysis. Bioorg. Med. Chem. Lett. 2009, 19, 4406–4409. [Google Scholar] [CrossRef]
- Brockhoff, A.; Behrens, M.; Massarotti, A.; Appendino, G.; Meyerhof, W. Broad tuning of the human bitter taste receptor hTAS2R46 to various sesquiterpene lactones, clerodane and labdane diterpenoids, strychnine, and denatonium. J. Agric. Food Chem. 2007, 55, 6236–6243. [Google Scholar] [CrossRef]
- Duffy, B.C.; Zhu, L.; Decornez, H.; Kitchen, D.B. Early phase drug discovery: Cheminformatics and computational techniques in identifying lead series. Bioorg. Med. Chem. 2012, 20, 5324–5342. [Google Scholar] [CrossRef]
- Heifetz, A.; Southey, M.; Morao, I.; Townsend-Nicholson, A.; Bodkin, M.J. Computational Methods Used in Hit-to-Lead and Lead Optimization Stages of Structure-Based Drug Discovery. Methods Mol. Biol. 2018, 1705, 375–394. [Google Scholar] [CrossRef]
- Fierro, F.; Suku, E.; Alfonso-Prieto, M.; Giorgetti, A.; Cichon, S.; Carloni, P. Agonist Binding to Chemosensory Receptors: A Systematic Bioinformatics Analysis. Front. Mol. Biosci. 2017, 4, 63. [Google Scholar] [CrossRef]
- Bahia, M.S.; Nissim, I.; Niv, M.Y. Bitterness prediction in-silico: A step towards better drugs. Int. J. Pharm. 2018, 536, 526–529. [Google Scholar] [CrossRef]
- Roland, W.S.; van Buren, L.; Gruppen, H.; Driesse, M.; Gouka, R.J.; Smit, G.; Vincken, J.P. Bitter Taste Receptor Activation by Flavonoids and Isoflavonoids: Modeled Structural Requirements for Activation of hTAS2R14 and hTAS2R39. J. Agric. Food Chem. 2013, 61, 10454–10466. [Google Scholar] [CrossRef]
- Levit, A.; Nowak, S.; Peters, M.; Wiener, A.; Meyerhof, W.; Behrens, M.; Niv, M.Y. The bitter pill: Clinical drugs that activate the human bitter taste receptor TAS2R14. FASEB J. 2013. [Google Scholar] [CrossRef]
- Ley, J.P.; Dessoy, M.; Paetz, S.; Blings, M.; Hoffmann-Lucke, P.; Reichelt, K.V.; Krammer, G.E.; Pienkny, S.; Brandt, W.; Wessjohann, L. Identification of Enterodiol as a Masker for Caffeine Bitterness by Using a Pharmacophore Model Based on Structural Analogues of Homoeriodictyol. J. Agric. Food Chem. 2012, 60, 6303–6311. [Google Scholar] [CrossRef]
- Liszt, K.I.; Ley, J.P.; Lieder, B.; Behrens, M.; Stoger, V.; Reiner, A.; Hochkogler, C.M.; Kock, E.; Marchiori, A.; Hans, J.; et al. Caffeine induces gastric acid secretion via bitter taste signaling in gastric parietal cells. Proc. Natl. Acad. Sci. USA 2017, 114, E6260–E6269. [Google Scholar] [CrossRef] [Green Version]
- Sanz, G.; Thomas-Danguin, T.; Hamdani, E.H.; Le Poupon, C.; Briand, L.; Pernollet, J.-C.; Guichard, E.; Tromelin, A. Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chem. Senses 2008, 33, 639–653. [Google Scholar] [CrossRef]
- Behrens, M.; Briand, L.; de March, C.A.; Matsunami, H.; Yamashita, A.; Meyerhof, W.; Weyand, S. Structure-Function Relationships of Olfactory and Taste Receptors. Chem. Senses 2018, 43, 81–87. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Fidelis, K.; Moult, J. CASP10 results compared to those of previous CASP experiments. Proteins 2013, 82 (Suppl. 2), 164–174. [Google Scholar] [CrossRef] [Green Version]
- Schneider, J.; Korshunova, K.; Musiani, F.; Alfonso-Prieto, M.; Giorgetti, A.; Carloni, P. Predicting ligand binding poses for low-resolution membrane protein models: Perspectives from multiscale simulations. Biochem. Biophys. Res. Commun. 2018, 498, 366–374. [Google Scholar] [CrossRef]
- Di Pizio, A.; Kruetzfeldt, L.M.; Cheled-Shoval, S.; Meyerhof, W.; Behrens, M.; Niv, M.Y. Ligand binding modes from low resolution GPCR models and mutagenesis: Chicken bitter taste receptor as a test-case. Sci. Rep. 2017, 7, 8223. [Google Scholar] [CrossRef]
- Morini, G.; Bassoli, A.; Temussi, P.A. From small sweeteners to sweet proteins: Anatomy of the binding sites of the human T1R2_T1R3 receptor. J. Med. Chem. 2005, 48, 5520–5529. [Google Scholar] [CrossRef]
- Zhang, F.; Klebansky, B.; Fine, R.M.; Xu, H.; Pronin, A.; Liu, H.; Tachdjian, C.; Li, X. Molecular mechanism for the umami taste synergism. Proc. Natl. Acad. Sci. USA 2008, 105, 20930–20934. [Google Scholar] [CrossRef] [Green Version]
- Dang, Y.L.; Hao, L.; Cao, J.X.; Sun, Y.Y.; Zeng, X.Q.; Wu, Z.; Pan, D.D. Molecular docking and simulation of the synergistic effect between umami peptides, monosodium glutamate and taste receptor T1R1/T1R3. Food Chem. 2019, 271, 697–706. [Google Scholar] [CrossRef]
- Kim, S.K.; Chen, Y.; Abrol, R.; Goddard, W.A.; Guthrie, B. Activation mechanism of the G protein-coupled sweet receptor heterodimer with sweeteners and allosteric agonists. Proc. Natl. Acad. Sci. USA 2017, 114, 2568–2573. [Google Scholar] [CrossRef]
- Zhang, F.; Klebansky, B.; Fine, R.M.; Liu, H.; Xu, H.; Servant, G.; Zoller, M.; Tachdjian, C.; Li, X. Molecular mechanism of the sweet taste enhancers. Proc. Natl. Acad. Sci. USA 2010, 107, 4752–4757. [Google Scholar] [CrossRef] [Green Version]
- Mouritsen, O.G.; Khandelia, H. Molecular mechanism of the allosteric enhancement of the umami taste sensation. FEBS J. 2012, 279, 3112–3120. [Google Scholar] [CrossRef] [Green Version]
- Wodak, S.J.; Paci, E.; Dokholyan, N.V.; Berezovsky, I.N.; Horovitz, A.; Li, J.; Hilser, V.J.; Bahar, I.; Karanicolas, J.; Stock, G.; et al. Allostery in Its Many Disguises: From Theory to Applications. Structure 2019. [Google Scholar] [CrossRef]
- Gelis, L.; Wolf, S.; Hatt, H.; Neuhaus, E.M.; Gerwert, K. Prediction of a Ligand-Binding Niche within a Human Olfactory Receptor by Combining Site-Directed Mutagenesis with Dynamic Homology Modeling. Angew. Chem. Int. Edit. 2012, 51, 1274–1278. [Google Scholar] [CrossRef]
- Geithe, C.; Protze, J.; Kreuchwig, F.; Krause, G.; Krautwurst, D. Structural determinants of a conserved enantiomer-selective carvone binding pocket in the human odorant receptor OR1A1. Cell. Mol. Life Sci. 2017, 74, 4209–4229. [Google Scholar] [CrossRef]
- Ahmed, L.; Zhang, Y.; Block, E.; Buehl, M.; Corr, M.J.; Cormanich, R.A.; Gundala, S.; Matsunami, H.; O’Hagan, D.; Ozbil, M.; et al. Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc. Natl. Acad. Sci. USA 2018, 115, E3950–E3958. [Google Scholar] [CrossRef]
- Cichero, E.; Espinoza, S.; Tonelli, M.; Franchini, S.; Gerasimov, A.S.; Sorbi, C.; Gainetdinov, R.R.; Brasili, L.; Fossa, P. A homology modelling-driven study leading to the discovery of the first mouse trace amine-associated receptor 5 (TAAR5) antagonists. Medchemcomm 2016, 7, 353–364. [Google Scholar] [CrossRef]
- Born, S.; Levit, A.; Niv, M.Y.; Meyerhof, W.; Behrens, M. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J. Neurosci. 2013, 33, 201–213. [Google Scholar] [CrossRef]
- Brockhoff, A.; Behrens, M.; Niv, M.Y.; Meyerhof, W. Structural requirements of bitter taste receptor activation. Proc. Natl. Acad. Sci. USA 2010, 107, 11110–11115. [Google Scholar] [CrossRef]
- Nowak, S.; Di Pizio, A.; Levit, A.; Niv, M.Y.; Meyerhof, W.; Behrens, M. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2162–2173. [Google Scholar] [CrossRef]
- Marchiori, A.; Capece, L.; Giorgetti, A.; Gasparini, P.; Behrens, M.; Carloni, P.; Meyerhof, W. Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: Experimentally-validated detailed structural prediction of agonist binding. PLoS ONE 2013, 8, e64675. [Google Scholar] [CrossRef]
- Jaggupilli, A.; Singh, N.; De Jesus, V.C.; Gounni, M.S.; Dhanaraj, P.; Chelikani, P. Chemosensory bitter taste receptors (T2Rs) are activated by multiple antibiotics. FASEB J. 2019, 33, 501–517. [Google Scholar] [CrossRef]
- Di Pizio, A.; Shy, N.; Behrens, M.; Meyerhof, W.; Niv, M.Y. Molecular Features Underlying Selectivity in Chicken Bitter Taste Receptors. Front. Mol. Biosci. 2018, 5, 6. [Google Scholar] [CrossRef]
- Sakurai, T.; Misaka, T.; Ishiguro, M.; Masuda, K.; Sugawara, T.; Ito, K.; Kobayashi, T.; Matsuo, S.; Ishimaru, Y.; Asakura, T.; et al. Characterization of the beta-D-Glucopyranoside Binding Site of the Human Bitter Taste Receptor hTAS2R16. J. Biol. Chem. 2010, 285, 28373–28378. [Google Scholar] [CrossRef]
- Sandal, M.; Behrens, M.; Brockhoff, A.; Musiani, F.; Giorgetti, A.; Carloni, P.; Meyerhof, W. Evidence for a Transient Additional Ligand Binding Site in the TAS2R46 Bitter Taste Receptor. J. Chem. Theory Comput. 2015, 11, 4439–4449. [Google Scholar] [CrossRef]
- Di Pizio, A.; Waterloo, L.A.W.; Brox, R.; Löber, S.; Weikert, D.; Behrens, M.; Gmeiner, P.; Niv, M.Y. Rational design of agonists for bitter taste receptor TAS2R14: From modeling to bench and back. Cell. Mol. Life Sci. 2019. under revision. [Google Scholar]




| MW | AlogP | HBAs | HBDs | ||
|---|---|---|---|---|---|
| Bitter tastants | Range | 27–1524 | −8.2–12.6 | 0–28 | 0–19 |
| Drug-like | 67% | 85% | 70% | 80% | |
| Sweet tastants a | Range | 92–1287 | −6.1–5.1 | 0–29 | 0–19 |
| Drug-like | 27% | 85% | 23% | 20% | |
| Umami tastants b | Range | 89–388 | −6.6–4.2 | 0–8 | 0–6 |
| Drug-like | 70% | 54% | 84% | 78% | |
| Odorants | Range | 44–345 | −1.2–4.2 | 0–4 | 0–2 |
| Drug-like | 4% | 99% | 100% | 100% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pizio, A.; Behrens, M.; Krautwurst, D. Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors. Int. J. Mol. Sci. 2019, 20, 1402. https://doi.org/10.3390/ijms20061402
Di Pizio A, Behrens M, Krautwurst D. Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors. International Journal of Molecular Sciences. 2019; 20(6):1402. https://doi.org/10.3390/ijms20061402
Chicago/Turabian StyleDi Pizio, Antonella, Maik Behrens, and Dietmar Krautwurst. 2019. "Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors" International Journal of Molecular Sciences 20, no. 6: 1402. https://doi.org/10.3390/ijms20061402
APA StyleDi Pizio, A., Behrens, M., & Krautwurst, D. (2019). Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors. International Journal of Molecular Sciences, 20(6), 1402. https://doi.org/10.3390/ijms20061402