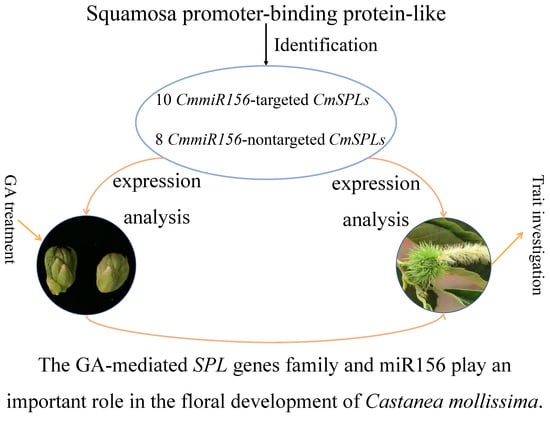
Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima)
Abstract
:1. Introduction
2. Results
2.1. Effects of GA and Its Inhibitor on the Number of Male and Female Inflorescences
2.2. Identification of CmSPL Genes
2.3. Phylogenetic Analysis of SPL Genes
2.4. Identification of Conserved Motifs in CmSPLs
2.5. Expression Profiles of the SPL Genes and miR156 in Organs of C. mollissima
2.6. Expression Pattern of miR156 and Its Target Genes in Floral Buds at Different Stages under GA Treatment
2.7. Validation of CmmiR156-Targeted CmSPLs Cleavage Sites by RNA Ligase-Mediated Rapid Amplification of the 5′ cDNA Ends (RLM-RACE)
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Exogenous Hormone Application
4.3. Identification of CmSPL Genes
4.4. Bioinformatic Analyses of CmSPL Genes
4.5. Phylogenetic Analysis
4.6. RNA Isolation
4.7. Expression Analysis of CmSPL Genes
4.8. Expression Analysis of CmmiR156
4.9. RLM-RACE
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Primer Set | |
|---|---|
| Forward and Reverse primer (5′-3′) | |
| For qRT-PCR analysis | |
| miRNA RT Primer | GATCGCCCTTCTACGTCGTATCGTCATCTGACCGTTATCGCTGCACGTTTTTTTTTTTTTTTTTTTT |
| CmmiR156-F | TGCACTAGCGTGTGACAGAAGA |
| CmmiR156-R | GATCGCCCTTCTACGTCGTAT |
| U6-F | GATAAAATTGGAACGATACAG |
| U6-R | ATTTGGACCATTTCTCGATTT |
| CmActin-F | ATTCACGAGACCACCTACA |
| CmActin-R | TGCCACAACCTTAATCTTCA |
| CmSPL1-F | CAGTCAAGTCCACCTCAA |
| CmSPL1-R | GCTCTCAATGTCAGTAGGA |
| CmSPL2-F | CATACCATAGGAGGCACAA |
| CmSPL2-R | TCCGTCGCTCATTATGTC |
| CmSPL3-F | TGTAGATGATGACTTGGATGA |
| CmSPL3-R | TTCTCTGCCTGACAACAT |
| CmSPL4-F | CAATGGAGGAACTAGGAGATG |
| CmSPL4-R | GACAATAATCCGAATGCTGAG |
| CmSPL5-F | TCTTCAAGCCATTCATCTG |
| CmSPL5-R | CCAAGGTGATTCTCTTCTG |
| CmSPL6-F | CTGAAGACTATGCTGTTACTG |
| CmSPL6-R | TGACCGACACTATGATGG |
| CmSPL7-F | GAACGCCACAACAATAGA |
| CmSPL7-R | TCAGACTCCAAGAATGCT |
| CmSPL8-F | ATGCTGGACTACGAATGG |
| CmSPL8-R | GATGATGATGTTGATGATGATGA |
| CmSPL9-F | TCCTCCATCTAACCTATATCTG |
| CmSPL9-R | CTTCCGTATTCATCAAGTCTT |
| CmSPL10-F | ACACCACGCTGCCAAGTT |
| CmSPL10-R | GGAACCTGCTACATTGCTGACA |
| CmSPL11-F | TACACCTCAAGTCCTTAGTCA |
| CmSPL11-R | GCTTCACTGCTTCGGTTA |
| CmSPL12-F | TTCTTCACAGTCACCATCT |
| CmSPL12-R | GCTCTCAATGTCAGTAGGA |
| CmSPL13-F | CTAACCAGCAGCAACAATTACAG |
| CmSPL13-R | AGACAGGAGCCTCAAGAGATAA |
| CmSPL14-F | GCAAGAAGTAGTAGCAGTTAC |
| CmSPL14-R | GGAGAAGACGAAGGAGAC |
| CmSPL15-F | TATCACCGCCGCCACAAG |
| CmSPL15-R | TTCCGTCGCCTCTCATTATGTC |
| CmSPL16-F | TTTATGCGACAATTACAAAGGA |
| CmSPL16-R | GTTTAACCCATTAGAGAACACTT |
| CmSPL17-F | TCTATCTTCTGTCATCACCTC |
| CmSPL17-R | TCATTGGCATCAGGAACA |
| CmSPL18-F | CACAAGCGTCACAAGGTT |
| CmSPL18-R | TCTCTGCCTGAAGGTCTG |
| For modified 5′ RLM-RACE | |
| CmSPL9-outer-R | GGGGAGGTGTCATAAGCCCTGGAGT |
| CmSPL9-inter-R | CCCTGGAGTGCTCAAGTCCCATGTA |
| CmSPL16-outer-R | ACAAGAAGGAAAGTGTTTGGTTAGG |
| CmSPL16-inter-R | TGACCCATCACTATCATGAGGCCCA |
References
- Zhao, Z.Y.; Liu, K.Y. Effect of chemical thinning catkins on Chinese chestnut yield and quality. Acta Hortic. 2009, 844, 457–460. [Google Scholar] [CrossRef]
- Guo, X.P.; Li, X.L.; Duan, X.W.; Shen, Y.Y.; Xing, Y.; Cao, Q.Q.; Qin, L. Characterization of sck1, a novel Castanea mollissima mutant with the extreme short catkins and decreased gibberellin. PLoS ONE 2012, 7, e43181. [Google Scholar] [CrossRef]
- Tanurdzic, M.; Banks, J.A. Sex-determining mechanisms in land plants. Plant Cell 2004, 16, S61–S71. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. The Hormonal Regulation of Flower Development. J. Plant Growth Regul. 2011, 30, 242–254. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Huang, H.; Xie, D. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol. Plant 2013, 6, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.; Ferguson, A.C.; Powers, S.J.; Wanchoo-Kohli, A.; Phillips, A.L.; Wilson, Z.A.; Hedden, P.; Thomas, S.G. DELLA activity is required for successful pollen development in the Columbia eco type of Arabidopsis. New Phytol. 2014, 201, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Zhao, Y.D. A Role for Auxin in Flower Development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.S.; Pan, B.Z.; Wang, G.J.; Ni, J.; Niu, L.; Xu, Z.F. Analysis of the transcriptional responses in inflorescence buds of Jatropha curcas exposed to cytokinin treatment. BMC Plant Biol. 2014, 14, 318. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.Z.; Xu, Z.F. Benzyladenine Treatment Significantly Increases the Seed Yield of the Biofuel Plant Jatropha curcas. J. Plant Growth Regul. 2011, 30, 166–174. [Google Scholar] [CrossRef]
- Fan, X.M.; Yuan, D.Y.; Tian, X.M.; Zhu, Z.J.; Liu, M.L.; Cao, H.P. Comprehensive Transcriptome Analysis of Phytohormone Biosynthesis and Signaling Genes in the Flowers of Chinese Chinquapin (Castanea henryi). J. Agric. Food Chem. 2017, 65, 10332–10349. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Pang, M.; Nah, G.; Shi, X.; Ye, W.; Stelly, D.M.; Chen, Z.J. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat. Commun. 2014, 5, 3050. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Z.Y.; Zhao, D.Y. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 2016, 6, 29938. [Google Scholar] [CrossRef]
- Tian, J.; Zhao, X.Y.; Xie, L.S.; Quan, J.Y.; Yao, L.H.; Wang, G.D.; Zheng, Y.Q.; Liu, X.M. Research advances and molecular mechanism on SPL transcription factors in regulating plant flower development. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2018, 42, 159–166. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef]
- Cardon, G.; Hohmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Li, C.L.; Lu, S.F. Molecular characterization of the SPL gene family in Populus trichocarpa. Biogeochemistry 2014, 14, 131. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Lin, E.P.; Huang, H.H.; Niu, M.Y.; Tong, Z.K.; Zhang, J.H. Molecular Characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family in Betula luminifera. Front. Plant Sci. 2018, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.T.; Hao, M.Y.; Wang, W.X.; Mei, D.S.; Tong, C.B.; Wang, H.; Liu, J.; Fu, L.; Hu, Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016, 16, 196. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, S.S.; Chen, F.; Liu, B.J.; Wu, L.; Li, F.; Zhang, J.P.; Bao, M.Z.; Liu, G.F. Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genom. 2018, 19, 1–18. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, S.P.; Salinas, M.; Höhmann, S.; Xing, S.; Berndtgen, R.; Huijser, P. miR156 Targeted and Nontargeted SBP-Box Transcription Factors Act in Concert to Secure Male Fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Kohalmi, S.E.; Amyot, L.; Hannoufa, A. SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Mol. Biol. 2016, 92, 661–674. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2014, 116, 281–297. [Google Scholar] [CrossRef]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.J.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, F.L.; Cao, H.; Peng, H.R.; Ni, Z.F.; Sun, Q.X.; Yao, Y.Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef]
- Li, J.; Guo, G.H.; Guo, W.W.; Guo, G.G.; Tong, D.; Ni, Z.F.; Sun, Q.F.; Yao, Y.Y. miR164-directedcleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol. 2012, 12, 220. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Duan, X.Y.; Zhang, Q.; Li, X.R.; Wang, B.; Huang, L.L.; Wang, X.J.; Kang, Z.S. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.F.F.E.; Silva, E.M.; Da Silva Azevedo, M.; Guivin, M.A.C.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.P.; Nogueira, F.T.S. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 2015, 8, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Long, J.M.; Liu, C.Y.; Feng, M.Q.; Liu, Y.; Wu, X.M.; Guo, W.W. miR156-SPLs module regulates somatic embryogenesis induction in citrus callus. J. Exp. Bot. 2018, 69, 2979–2993. [Google Scholar] [CrossRef]
- Gou, J.Q.; Fu, C.X.; Liu, S.J.; Tang, C.R.; Debnath, S.; Flanagan, A.; Ge, Y.X. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017, 216, 829. [Google Scholar] [CrossRef]
- Shang, M.Y.; Wang, X.T.; Zhang, J.; Qi, X.H.; Ping, A.; Hou, L.P.; Xing, G.M.; Li, G.Z.; Li, M.L. Genetic Regulation of GA Metabolism during Vernalization, Floral Bud Initiation and Development in Pak Choi (Brassica rapa ssp. chinensis Makino). Front. Plant Sci. 2017, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wen, J.; Zimmer, E.A. Expression patterns of AP1, FUL, FT and LEAFY orthologs in Vitaceae support the homology of tendrils and inflorescences throughout the grape family. J. Syst. Evol. 2015, 53, 469–476. [Google Scholar] [CrossRef]
- Dong, B.; Deng, Y.; Wang, H.B.; Gao, R.; Stephen, G.K.; Chen, S.M.; Jiang, J.F.; Chen, F.D. Gibberellic Acid Signaling Is Required to Induce Flowering of Chrysanthemums Grown under Both Short and Long Days. Int. J. Mol. Sci. 2017, 18, 1259. [Google Scholar] [CrossRef]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Galvão, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.W. Gibberellin regulates the Arabidopsis floral transition through miR156 targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Liu, Z.J.; Wang, L.G.; Sheng, Z.; Xie, J.P.; Bi, Y.R. Sucrose-induced hypocotyl elongation of Arabidopsis seedlings in darkness depends on the presence of gibberellins. J. Plant Physiol. 2010, 167, 1130–1136. [Google Scholar] [CrossRef]
- Boccaccini, A.; Santopolo, S.; Capauto, D.; Lorrai, R.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. The DOF protein DAG1 and the DELLA protein GAI cooperate in negatively regulating the AtGA3ox1 gene. Mol. Plant 2017, 7, 1486–1489. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Gao, Z.P.; Zhu, Z.Q. DELLA-PIF modules: Old dogs learn new tricks. Trends Plant Sci. 2016, 21, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Wang, Y.; Liu, H.L.; Wu, M.; Chu, W.Y.; Chen, D.M.; Xiang, Y. Genome-wide identification and expression analysis of SBP-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). BMC Genom. 2017, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.P.; Guo, W.Z.; Zhang, B.H. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 2018, 8, 762. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Wei, W.; Zhou, W.; McGuigan, L.D.; Ji, F.Y.; Li, X.; Xing, Y.; Zhang, Q.; Fang, K.F.; Cao, Q.Q. Establishment of a somatic embryo regeneration system and expression analysis of somatic embryogenesis-related genes in Chinese chestnut (Castanea mollissima Blume). Plant Cell Tissue Organ Cult. 2017, 130, 601–616. [Google Scholar] [CrossRef]







| Species | Concentration (mg·L−1) | Female Flower Cluster /Branch | Number of Mixed Inflorescences /Branch | Mixed Inflorescence Length (cm) | Number of Male Inflorescence /Branch | Ratio of Female Flower Clusters to Mixed Inflorescence Number | The Ratio of the Number of Female Flowers to the Number of Male Inflorescences | Ratio of Mixed Inflorescence Number to Male Inflorescence Number |
|---|---|---|---|---|---|---|---|---|
| C | 0 | 2.71 ± 1.62b | 2.11 ± 0.93a | 16.33 ± 2.61a | 8.44 ± 3.43b | 1.5 ± 0.40a | 0.47 ± 0.27a | 0.34 ± 0.15a |
| GA | 100 mg/L | 2.36 ± 0.50b | 2.14 ± 0.53a | 12.65 ± 2.34b | 11.14 ± 2.98a | 1.14 ± 0.31b | 0.22 ± 0.06b | 0.21 ± 0.07a |
| PP333 | 1000 mg/L | 3.70 ± 1.42a | 2.40 ± 0.84a | 17.40 ± 2.94a | 8.80 ± 1.87b | 1.58 ± 0.34a | 0.46 ± 0.25a | 0.30 ± 0.15a |
| Gene Name | CDS (bp) | Peptide (aa) | Mw (Da) | pI | Ai | miR156 Target |
|---|---|---|---|---|---|---|
| CmSPL1 | 3000 | 999 | 110899.70 | 5.77 | 84.41 | No |
| CmSPL2 | 582 | 193 | 21864.54 | 9.39 | 51.50 | Yes |
| CmSPL3 | 549 | 182 | 20368.72 | 8.73 | 51.43 | No |
| CmSPL4 | 1314 | 437 | 48311.02 | 7.05 | 57.41 | Yes |
| CmSPL5 | 1722 | 573 | 62793.15 | 6.66 | 57.87 | Yes |
| CmSPL6 | 1623 | 540 | 60015.14 | 6.15 | 68.24 | Yes |
| CmSPL7 | 2358 | 785 | 87807.21 | 5.72 | 76.28 | No |
| CmSPL8 | 1026 | 341 | 37999.46 | 8.98 | 44.75 | No |
| CmSPL9 | 1119 | 372 | 39809.28 | 9.37 | 47.55 | Yes |
| CmSPL10 | 1410 | 469 | 52259.35 | 8.65 | 52.47 | Yes |
| CmSPL11 | 1398 | 465 | 51284.57 | 8.03 | 58.11 | Yes |
| CmSPL12 | 3108 | 1035 | 114576.19 | 7.32 | 84.38 | No |
| CmSPL13 | 1176 | 391 | 42863.75 | 8.80 | 62.58 | Yes |
| CmSPL14 | 3276 | 1091 | 120636.02 | 7.86 | 74.28 | No |
| CmSPL15 | 456 | 151 | 17311.85 | 6.71 | 29.74 | No |
| CmSPL16 | 960 | 319 | 34804.61 | 9.15 | 60.19 | Yes |
| CmSPL17 | 1173 | 390 | 43054.10 | 8.47 | 69.00 | Yes |
| CmSPL18 | 624 | 207 | 23511.61 | 9.32 | 53.24 | No |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Li, J.; Liu, Y.; Zhang, Q.; Gao, Y.; Fang, K.; Cao, Q.; Qin, L.; Xing, Y. Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima). Int. J. Mol. Sci. 2019, 20, 1577. https://doi.org/10.3390/ijms20071577
Chen G, Li J, Liu Y, Zhang Q, Gao Y, Fang K, Cao Q, Qin L, Xing Y. Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima). International Journal of Molecular Sciences. 2019; 20(7):1577. https://doi.org/10.3390/ijms20071577
Chicago/Turabian StyleChen, Guosong, Jingtong Li, Yang Liu, Qing Zhang, Yuerong Gao, Kefeng Fang, Qingqin Cao, Ling Qin, and Yu Xing. 2019. "Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima)" International Journal of Molecular Sciences 20, no. 7: 1577. https://doi.org/10.3390/ijms20071577
APA StyleChen, G., Li, J., Liu, Y., Zhang, Q., Gao, Y., Fang, K., Cao, Q., Qin, L., & Xing, Y. (2019). Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima). International Journal of Molecular Sciences, 20(7), 1577. https://doi.org/10.3390/ijms20071577