Rab39a and Rab39b Display Different Intracellular Distribution and Function in Sphingolipids and Phospholipids Transport
Abstract
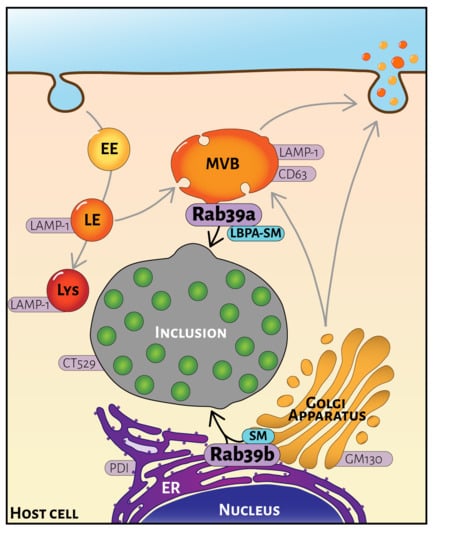
:1. Introduction
2. Results
2.1. Rab39a and Rab39b Intracellular Distribution
2.2. Motility of Rab39a- and Rab39b-Vesicles Along Microtubules
2.3. Dynamics of Rab39a- and Rab39b-Vesicles
2.4. Rab39a- and Rab39b-Mediated Lipid Transport
3. Discussion
4. Materials and Methods
4.1. Cells and Bacteria
4.2. Antibodies and Reagents
4.3. Plasmids and Cell Transfection
4.4. Immunofluorescence and Confocal Microscopy
4.5. Time-Lapse Video Microscopy
4.6. Vesicular Movement Analysis
4.7. Image Analysis
4.8. Staining of Lipids and Organelles
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef]
- Chen, T.; Han, Y.; Yang, M.; Zhang, W.; Li, N.; Wan, T.; Guo, J.; Cao, X. Rab39, a novel Golgi-associated Rab GTPase from human dendritic cells involved in cellular endocytosis. Biochem. Biophys. Res. Commun. 2003, 303, 1114–1120. [Google Scholar] [CrossRef]
- Seto, S.; Tsujimura, K.; Koide, Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic 2011, 12, 407–420. [Google Scholar] [CrossRef]
- Becker, C.E.; Creagh, E.M.; O’Neill, L.A. Rab39a binds caspase-1 and is required for caspase-1-dependent interleukin-1beta secretion. J. Biol. Chem. 2009, 284, 34531–34537. [Google Scholar] [CrossRef]
- Cheng, H.; Ma, Y.; Ni, X.; Jiang, M.; Guo, L.; Ying, K.; Xie, Y.; Mao, Y. Isolation and characterization of a human novel RAB (RAB39B) gene. Cytogenet. Genome Res. 2002, 97, 72–75. [Google Scholar] [CrossRef]
- Yuan, L.; Deng, X.; Song, Z.; Yang, Z.; Ni, B.; Chen, Y.; Deng, H. Genetic analysis of the RAB39B gene in Chinese Han patients with Parkinson’s disease. Neurobiol. Aging 2015, 36, 2907.e11-2. [Google Scholar] [CrossRef]
- Vanmarsenille, L.; Giannandrea, M.; Fieremans, N.; Verbeeck, J.; Belet, S.; Raynaud, M.; Vogels, A.; Männik, K.; Õunap, K.; Jacqueline, V.; et al. Increased dosage of RAB39B affects neuronal development and could explain the cognitive impairment in male patients with distal Xq28 copy number gains. Hum. Mutat. 2014, 35, 377–383. [Google Scholar] [CrossRef]
- Wilson, G.R.; Sim, J.C.H.; McLean, C.; Giannandrea, M.; Galea, C.A.; Riseley, J.R.; Stephenson, S.E.M.; Fitzpatrick, E.; Haas, S.A.; Pope, K.; et al. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with α-synuclein pathology. Am. J. Hum. Genet. 2014, 95, 729–735. [Google Scholar] [CrossRef]
- Corbier, C.; Sellier, C. C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases 2017, 8, 181–186. [Google Scholar] [CrossRef]
- Mignogna, M.L.; Giannandrea, M.; Gurgone, A.; Fanelli, F.; Raimondi, F.; Mapelli, L.; Bassani, S.; Fang, H.; Van Anken, E.; Alessio, M.; et al. The intellectual disability protein RAB39B selectively regulates GluA2 trafficking to determine synaptic AMPAR composition. Nat. Commun. 2015, 6, 6504. [Google Scholar] [CrossRef]
- Yao, Y.; Cui, X.; Al-Ramahi, I.; Sun, X.; Li, B.; Hou, J.; Difiglia, M.; Palacino, J.; Wu, Z.-Y.; Ma, L.; et al. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. eLife 2015. [Google Scholar] [CrossRef]
- Yoshimura, S.; Gerondopoulos, A.; Linford, A.; Rigden, D.J.; Barr, F.A. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 2010, 191, 367–381. [Google Scholar] [CrossRef] [Green Version]
- Proikas-Cezanne, T.; Gaugel, A.; Frickey, T.; Nordheim, A. Rab14 is part of the early endosomal clathrin-coated TGN microdomain. FEBS Lett. 2006, 580, 5241–5246. [Google Scholar] [CrossRef]
- Mueller, K.E.; Plano, G.V.; Fields, K.A. New frontiers in type III secretion biology: The Chlamydia perspective. Infect. Immun. 2014, 82, 2–9. [Google Scholar] [CrossRef]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef] [Green Version]
- Elwell, C.A.; Engel, J.N. Lipid acquisition by intracellular Chlamydiae. Cell. Microbiol. 2012, 14, 1010–1018. [Google Scholar] [CrossRef]
- Saka, H.A.; Valdivia, R.H. Acquisition of nutrients by Chlamydiae: Unique challenges of living in an intracellular compartment. Curr. Opin. Microbiol. 2010, 13, 4–10. [Google Scholar] [CrossRef]
- Damiani, M.T.; Gambarte Tudela, J.; Capmany, A. Targeting eukaryotic Rab proteins: A smart strategy for chlamydial survival and replication. Cell. Microbiol. 2014, 16, 1329–1338. [Google Scholar] [CrossRef]
- Hackstadt, T. Redirection of host vesicle trafficking pathways by intracellular parasites. Traffic 2000, 1, 93–99. [Google Scholar] [CrossRef]
- Gambarte Tudela, J.; Capmany, A.; Romao, M.; Quintero, C.; Miserey-Lenkei, S.; Raposo, G.; Goud, B.; Damiani, M.T. The late endocytic Rab39a GTPase regulates the interaction between multivesicular bodies and chlamydial inclusions. J. Cell Sci. 2015, 128, 3068–3081. [Google Scholar] [CrossRef] [Green Version]
- Rzomp, K.A.; Moorhead, A.R.; Scidmore, M.A. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 2006, 74, 5362–5373. [Google Scholar] [CrossRef]
- Leiva, N.; Capmany, A.; Damiani, M.T. Rab11-family of interacting protein 2 associates with chlamydial inclusions through its Rab-binding domain and promotes bacterial multiplication. Cell. Microbiol. 2013, 15, 114–129. [Google Scholar] [CrossRef]
- Rzomp, K.A.; Scholtes, L.D.; Briggs, B.J.; Whittaker, G.R.; Scidmore, M.A. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 2003, 71, 5855–5870. [Google Scholar] [CrossRef]
- Rejman Lipinski, A.; Heymann, J.; Meissner, C.; Karlas, A.; Brinkmann, V.; Meyer, T.F.; Heuer, D. Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 2009, 5, e1000615. [Google Scholar] [CrossRef]
- Capmany, A.; Damiani, M.T. Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a Rab14-mediated transport required for bacterial development and replication. PLoS ONE 2010, 5, e14084. [Google Scholar] [CrossRef]
- Lindsay, A.J.; Jollivet, F.; Horgan, C.P.; Khan, A.R.; Raposo, G.; McCaffrey, M.W.; Goud, B. Identification and characterization of multiple novel Rab–myosin Va interactions. Mol. Biol. Cell 2013, 24, 3420–3434. [Google Scholar] [CrossRef] [Green Version]
- Piper, R.C.; Katzmann, D.J. Biogenesis and Function of Multivesicular Bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [Green Version]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A Common Pathway for a Specialized Function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Kjos, I.; Vestre, K.; Guadagno, N.A.; Borg Distefano, M.; Progida, C. Rab and Arf proteins at the crossroad between membrane transport and cytoskeleton dynamics. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.D.; Stephens, D.J. The role of motor proteins in endosomal sorting. Biochem. Soc. Trans. 2011, 39, 1179–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delevoye, C.; Goud, B. Rab GTPases and kinesin motors in endosomal trafficking. Methods Cell Biol. 2015, 130, 235–246. [Google Scholar]
- Horgan, C.P.; McCaffrey, M.W. Rab GTPases and microtubule motors. Biochem. Soc. Trans. 2011, 39, 1202–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, B.; Costantino, S.; Wiseman, P.W. Spatiotemporal Image Correlation Spectroscopy (STICS) Theory, Verification, and Application to Protein Velocity Mapping in Living CHO Cells. Biophys. J. 2005, 88, 3601–3614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapierre, L.A.; Dorn, M.C.; Zimmerman, C.F.; Navarre, J.; Burnette, J.O.; Goldenring, J.R. Rab11b resides in a vesicular compartment distinct from Rab11a in parietal cells and other epithelial cells. Exp. Cell Res. 2003, 290, 322–331. [Google Scholar] [CrossRef]
- Bucci, C.; Lütcke, A.; Steele-Mortimer, O.; Olkkonen, V.M.; Dupree, P.; Chiariello, M.; Bruni, C.B.; Simons, K.; Zerial, M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 1995, 366, 65–71. [Google Scholar] [CrossRef]
- Chen, P.-I.; Kong, C.; Su, X.; Stahl, P.D. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J. Biol. Chem. 2009, 284, 30328–30338. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M. Rab27 effectors, pleiotropic regulators in secretory pathways. Traffic 2013, 14, 949–963. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Kobayashi, S.; Suzuki, T.; Kawaguchi, A.; Phongphaew, W.; Yoshii, K.; Iwano, T.; Harada, A.; Kariwa, H.; Orba, Y.; Sawa, H. Rab8b Regulates Transport of West Nile Virus Particles from Recycling Endosomes. J. Biol. Chem. 2016, 291, 6559–6568. [Google Scholar] [CrossRef]
- Carabeo, R.A.; Mead, D.J.; Hackstadt, T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. USA 2003, 100, 6771–6776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ZL, C.; Kumar, Y.; Fischer, E.R.; Hackstadt, T.; Valdivia, R.H. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. USA 2008, 105, 9379–9384. [Google Scholar] [Green Version]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for Cell and Particle Tracking. Methods Enzymol. 2012, 504, 183–200. [Google Scholar] [PubMed]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambarte Tudela, J.; Buonfigli, J.; Luján, A.; Alonso Bivou, M.; Cebrián, I.; Capmany, A.; Damiani, M.T. Rab39a and Rab39b Display Different Intracellular Distribution and Function in Sphingolipids and Phospholipids Transport. Int. J. Mol. Sci. 2019, 20, 1688. https://doi.org/10.3390/ijms20071688
Gambarte Tudela J, Buonfigli J, Luján A, Alonso Bivou M, Cebrián I, Capmany A, Damiani MT. Rab39a and Rab39b Display Different Intracellular Distribution and Function in Sphingolipids and Phospholipids Transport. International Journal of Molecular Sciences. 2019; 20(7):1688. https://doi.org/10.3390/ijms20071688
Chicago/Turabian StyleGambarte Tudela, Julián, Julio Buonfigli, Agustín Luján, Mariano Alonso Bivou, Ignacio Cebrián, Anahí Capmany, and María Teresa Damiani. 2019. "Rab39a and Rab39b Display Different Intracellular Distribution and Function in Sphingolipids and Phospholipids Transport" International Journal of Molecular Sciences 20, no. 7: 1688. https://doi.org/10.3390/ijms20071688
APA StyleGambarte Tudela, J., Buonfigli, J., Luján, A., Alonso Bivou, M., Cebrián, I., Capmany, A., & Damiani, M. T. (2019). Rab39a and Rab39b Display Different Intracellular Distribution and Function in Sphingolipids and Phospholipids Transport. International Journal of Molecular Sciences, 20(7), 1688. https://doi.org/10.3390/ijms20071688