DNA Methylation Contributes to the Differential Expression Levels of Mecp2 in Male Mice Neurons and Astrocytes
Abstract
:1. Introduction
2. Results
2.1. Establishment of Sex-Specific Cultures of Male and Female Primary Neurons and Astrocytes
2.2. Mecp2 Isoforms Show Cell Type- and Sex-Specific Expression in Neurons and Astrocytes
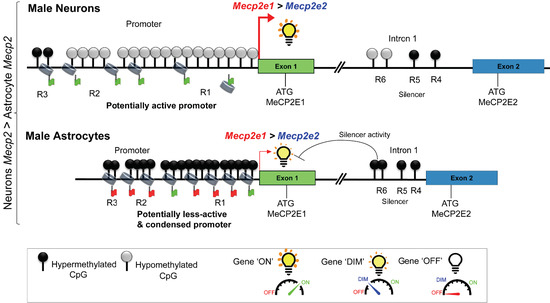
2.3. DNA Methylation at the Mecp2 Regulatory Elements May Contribute to Higher Expression of Mecp2 in Male Neurons Compared to Male Astrocytes with Lower Mecp2 Expression
2.4. Expression of Mecp2 Isoforms Correlates with DNA Methylation at the Mecp2 REs
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Primary Culture of Embryonic E18.5 Neurons
4.3. Primary Culture of Embryonic E18.5 Astrocytes
4.4. Culture and Identification of Sex-Specific Neurons and Astrocytes
4.5. Quantitative RT-PCR (qRT-PCR)
4.6. DNA Methylation Analysis by Bisulfite Pyrosequencing
4.7. Correlation Analysis between Detected DNA Methylation at the Mecp2 Regulatory Elements and Transcript Expression Levels of Mecp2 Isoforms
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5hmC | 5-hydrocymethylcytosine |
| 5mC | 5-methylcytosine |
| ASD | autism spectrum disorder |
| BIRC4 | baculoviral IAP repeat-containing protein 4 |
| bp | base pairs |
| cDNA | complementary DNA |
| CTCF | CCCTC-binding factor |
| DNMT | DNA methyltransferase |
| E | embryonic day |
| ENCODE | Encyclopedia of DNA Elements |
| F | female |
| FASD | fetal alcohol spectrum disorder |
| FBS | fetal bovine serum |
| GAB3 | GRB2-associated-binding protein 3 |
| Gapdh | glyceraldehyde 3-phosphate dehydrogenase |
| h | hours |
| Hdac | Histone deacetylase 6 |
| Il3 | interleukin 3 |
| JARID1C | Jumonji/ARID domain-containing protein 1C |
| Kdm6a | lysine demethylase 6A |
| M | male |
| MDS | MECP2 duplication syndrome |
| MeCP2 | Methyl CpG binding protein 2 |
| P | postnatal day |
| PCR | polymerase chain reaction |
| PolII-S5p | RNA polymerase phosphorylated at Ser 5 |
| qRT-PCR | quantitative reverse transcription PCR |
| R | region |
| r | Pearson’s correlation coefficient |
| REs | regulatory elements |
| RPS4X | ribosomal protein S4 X-linked |
| RTT | Rett syndrome |
| SEM | standard error of the mean |
| SLC16A2 | solute carrier family 16 member 2 |
| Sry | sex-determining region protein gene on the Y chromosome |
| UBE1 | ubiquitin-like modifier activating enzyme 1 |
| XCI | X chromosome inactivation |
| Xist | X-inactive specific transcript |
References
- Zachariah, R.M.; Rastegar, M. Linking epigenetics to human disease and Rett syndrome: The emerging novel and challenging concepts in MeCP2 research. Neural Plast. 2012, 2012, 415825. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Lee, S.S.; Zhang, X.; Houwink-Manville, I.; Song, H.R.; Amir, R.E.; Budden, S.; Naidu, S.; Pereira, J.L.; Lo, I.F.; et al. Rett syndrome and beyond: Recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am. J. Hum. Genet. 1999, 65, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Olson, C.O.; Pejhan, S.; Kroft, D.; Sheikholeslami, K.; Fuss, D.; Buist, M.; Ali Sher, A.; Del Bigio, M.R.; Sztainberg, Y.; Siu, V.M.; et al. MECP2 Mutation Interrupts Nucleolin-mTOR-P70S6K Signaling in Rett Syndrome Patients. Front. Genet. 2018, 9, 635. [Google Scholar] [CrossRef]
- Meins, M.; Lehmann, J.; Gerresheim, F.; Herchenbach, J.; Hagedorn, M.; Hameister, K.; Epplen, J.T. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. J. Med. Genet. 2005, 42, e12. [Google Scholar] [CrossRef]
- Ariani, F.; Mari, F.; Pescucci, C.; Longo, I.; Bruttini, M.; Meloni, I.; Hayek, G.; Rocchi, R.; Zappella, M.; Renieri, A. Real-time quantitative PCR as a routine method for screening large rearrangements in Rett syndrome: Report of one case of MECP2 deletion and one case of MECP2 duplication. Hum. Mutat. 2004, 24, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Van Esch, H. MECP2 Duplication Syndrome. In GeneReviews(R); Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Fong, C.T., Mefford, H.C., Smith, R.J.H., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Nagarajan, R.P.; Hogart, A.R.; Gwye, Y.; Martin, M.R.; LaSalle, J.M. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics 2006, 1, 172–182. [Google Scholar] [CrossRef]
- Nagarajan, R.P.; Patzel, K.A.; Martin, M.; Yasui, D.H.; Swanberg, S.E.; Hertz-Picciotto, I.; Hansen, R.L.; Van de Water, J.; Pessah, I.N.; Jiang, R.; et al. MECP2 promoter methylation and X chromosome inactivation in autism. Autism Res. 2008, 1, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, V.R.; Jarmasz, J.S.; Murugeshan, N.; Del Bigio, M.R.; Rastegar, M.; Davie, J.R. DNA modifications: Function and applications in normal and disease States. Boilogy 2014, 3, 670–723. [Google Scholar] [CrossRef]
- Liyanage, V.R.; Curtis, K.; Zachariah, R.M.; Chudley, A.E.; Rastegar, M. Overview of the Genetic Basis and Epigenetic Mechanisms that Contribute to FASD Pathobiology. Curr. Top. Med. Chem. 2017, 17, 808–828. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.C.; Choi, C.S.; Kim, J.W.; Han, S.H.; Cheong, J.H.; Ryu, J.H.; Shin, C.Y. MeCP2 Modulates Sex Differences in the Postsynaptic Development of the Valproate Animal Model of Autism. Mol. Neurobiol. 2016, 53, 40–56. [Google Scholar] [CrossRef]
- Deng, X.; Berletch, J.B.; Nguyen, D.K.; Disteche, C.M. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat. Rev. Genet. 2014, 15, 367–378. [Google Scholar] [CrossRef]
- Lioy, D.T.; Garg, S.K.; Monaghan, C.E.; Raber, J.; Foust, K.D.; Kaspar, B.K.; Hirrlinger, P.G.; Kirchhoff, F.; Bissonnette, J.M.; Ballas, N. A role for glia in the progression of Rett’s syndrome. Nature 2011, 475, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Zachariah, R.M.; Olson, C.O.; Ezeonwuka, C.; Rastegar, M. Novel MeCP2 isoform-specific antibody reveals the endogenous MeCP2E1 expression in murine brain, primary neurons and astrocytes. PLoS ONE 2012, 7, e49763. [Google Scholar] [CrossRef] [PubMed]
- Ballas, N.; Lioy, D.T.; Grunseich, C.; Mandel, G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 2009, 12, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Liyanage, V.R.; Zachariah, R.M.; Rastegar, M. Decitabine alters the expression of Mecp2 isoforms via dynamic DNA methylation at the Mecp2 regulatory elements in neural stem cells. Mol. Autism 2013, 4, 46. [Google Scholar] [CrossRef]
- Liyanage, V.R.; Zachariah, R.M.; Davie, J.R.; Rastegar, M. Ethanol deregulates Mecp2/MeCP2 in differentiating neural stem cells via interplay between 5-methylcytosine and 5-hydroxymethylcytosine at the Mecp2 regulatory elements. Exp. Neurol. 2015, 265, 102–117. [Google Scholar] [CrossRef] [Green Version]
- Olson, C.O.; Zachariah, R.M.; Ezeonwuka, C.D.; Liyanage, V.R.; Rastegar, M. Brain region-specific expression of MeCP2 isoforms correlates with DNA methylation within Mecp2 regulatory elements. PLoS ONE 2014, 9, e90645. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Liyanage, V.R.B.; MacAulay, A.; Levy, R.D.; Curtis, K.; Olson, C.O.; Zachariah, R.M.; Amiri, S.; Buist, M.; Hicks, G.G.; et al. Genome-Wide Transcriptome Landscape of Embryonic Brain-Derived Neural Stem Cells Exposed to Alcohol with Strain-Specific Cross-Examination in BL6 and CD1 Mice. Sci. Rep. 2019, 9, 206. [Google Scholar] [CrossRef]
- Loomes, R.; Hull, L.; Mandy, W.P.L. What Is the Male-to-Female Ratio in Autism Spectrum Disorder? A Systematic Review and Meta-Analysis. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fombonne, E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009, 65, 591–598. [Google Scholar] [CrossRef]
- Rastegar, M.; Hotta, A.; Pasceri, P.; Makarem, M.; Cheung, A.Y.; Elliott, S.; Park, K.J.; Adachi, M.; Jones, F.S.; Clarke, I.D.; et al. MECP2 isoform-specific vectors with regulated expression for Rett syndrome gene therapy. PLoS ONE 2009, 4, e6810. [Google Scholar] [CrossRef]
- Barber, B.A.; Liyanage, V.R.; Zachariah, R.M.; Olson, C.O.; Bailey, M.A.; Rastegar, M. Dynamic expression of MEIS1 homeoprotein in E14.5 forebrain and differentiated forebrain-derived neural stem cells. Ann. Anat. 2013, 195, 431–440. [Google Scholar] [CrossRef]
- Cerase, A.; Pintacuda, G.; Tattermusch, A.; Avner, P. Xist localization and function: New insights from multiple levels. Genome Boil. 2015, 16, 166. [Google Scholar] [CrossRef]
- Dewing, P.; Chiang, C.W.; Sinchak, K.; Sim, H.; Fernagut, P.O.; Kelly, S.; Chesselet, M.F.; Micevych, P.E.; Albrecht, K.H.; Harley, V.R.; et al. Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 2006, 16, 415–420. [Google Scholar] [CrossRef]
- Lambert, J.-F.; Benoit, B.O.; Colvin, G.A.; Carlson, J.; Delville, Y.; Quesenberry, P.J. Quick sex determination of mouse fetuses. J. Neurosci. Methods 2000, 95, 127–132. [Google Scholar] [CrossRef]
- Patrat, C.; Okamoto, I.; Diabangouaya, P.; Vialon, V.; Le Baccon, P.; Chow, J.; Heard, E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl. Acad. Sci. USA 2009, 106, 5198–5203. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Tan, P.P.C.; Portales-Casamar, E.; Pavlidis, P. Meta-analysis of human methylomes reveals stably methylated sequences surrounding CpG islands associated with high gene expression. Epigenet. Chromatin 2014, 7, 28. [Google Scholar] [CrossRef]
- Doi, A.; Park, I.-H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S. Differential methylation of tissue-and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Van Veldhoven, K.; Polidoro, S.; Baglietto, L.; Severi, G.; Sacerdote, C.; Panico, S.; Mattiello, A.; Palli, D.; Masala, G.; Krogh, V.; et al. Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin. Epigenet. 2015, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.D.; Timp, W.; Bravo, H.C.; Sabunciyan, S.; Langmead, B.; McDonald, O.G.; Wen, B.; Wu, H.; Liu, Y.; Diep, D. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 2011, 43, 768–775. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.-Y.; Hsu, S.-D.; Huang, H.-Y.; Sun, Y.-M.; Chou, C.-H.; Weng, S.-L.; Huang, H.-D. MethHC: A database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2014, 43, D856–D861. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Hsu, S.D.; Huang, H.Y.; Sun, Y.M.; Chou, C.H.; Weng, S.L.; Huang, H.D. MethHC: A Database of DNA Methylation and Gene Expression in Human Cancer. Available online: http://methhc.mbc.nctu.edu.tw/php/index.php (accessed on 6 February 2017).
- Vieira, J.P.; Lopes, F.; Silva-Fernandes, A.; Sousa, M.V.; Moura, S.; Sousa, S.; Costa, B.M.; Barbosa, M.; Ylstra, B.; Temudo, T.; et al. Variant Rett syndrome in a girl with a pericentric X-chromosome inversion leading to epigenetic changes and overexpression of the MECP2 gene. Int. J. Dev. Neurosci. 2015, 46, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delcuve, G.P.; Rastegar, M.; Davie, J.R. Epigenetic control. J. Cell. Physiol. 2009, 219, 243–250. [Google Scholar] [CrossRef] [Green Version]
- Kurian, J.R.; Forbes-Lorman, R.M.; Auger, A.P. Sex Difference in Mecp2 Expression During a Critical Period of Rat Brain Development. Epigenetics 2007, 2, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berletch, J.B.; Ma, W.; Yang, F.; Shendure, J.; Noble, W.S.; Disteche, C.M.; Deng, X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015, 11, e1005079. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Driscoll, D.J. Trimethylation of histone H3 lysine 4 is an epigenetic mark at regions escaping mammalian X inactivation. Epigenetics 2007, 2, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Nino-Soto, M.; Nuber, U.A.; Basrur, P.; Ropers, H.-H.; King, W. Differences in the pattern of X-linked gene expression between fetal bovine muscle and fibroblast cultures derived from the same muscle biopsies. Cytogenet. Genome Res. 2005, 111, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Talebizadeh, Z.; Simon, S.D.; Butler, M.G. X chromosome gene expression in human tissues: Male and female comparisons. Genomics 2006, 88, 675–681. [Google Scholar] [CrossRef]
- Rastegar, M. Epigenetics and Cerebellar Neurodevelopmental Disorders. In Development of the Cerebellum from Molecular Aspects to Diseases; Marzban, H., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 197–218. [Google Scholar]
- Marzban, H.; Del Bigio, M.R.; Alizadeh, J.; Ghavami, S.; Zachariah, R.M.; Rastegar, M. Cellular commitment in the developing cerebellum. Front. Cell. Neurosci. 2015, 8, 450. [Google Scholar] [CrossRef]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Pastor, W.A.; Shen, Y.; Tahiliani, M.; Liu, D.R.; Rao, A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 2010, 5, e8888. [Google Scholar] [CrossRef]
- Qui, Y.; Yang, Q.; Sui, F.; Lu, R.; Dang, S.; Ji, M.; He, N.; Shi, B.; Hou, P. A Strategy for Accurate Quantification of 5-Methylcytosine and 5-Hydroxymethylcytosine at CpG Sites Within Gene Promoter. J. Biomed. Nanotechnol. 2015, 11, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Yu, D.; Sakurai, F.; Corey, D.R. Clonal Rett Syndrome cell lines to test compounds for activation of wild-type MeCP2 expression. Bioorg. Med. Chem. Lett. 2011, 21, 5202–5205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, Y.; Sun, X.-H. Zebularine suppresses TGF-beta-induced lens epithelial cell–myofibroblast transdifferentiation by inhibiting MeCP2. Molecular 2011, 17, 2717. [Google Scholar]
- He, S.; Barron, E.; Ishikawa, K.; Khanamiri, H.N.; Spee, C.; Zhou, P.; Kase, S.; Wang, Z.; Dustin, L.D.; Hinton, D.R. Inhibition of DNA Methylation and Methyl-CpG-Binding Protein 2 Suppresses RPE Transdifferentiation: Relevance to Proliferative VitreoretinopathyDNA Methylation Regulates RPE Transdifferentiation. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5579–5589. [Google Scholar] [CrossRef]
- Hartshorn, C.; Rice, J.E.; Wangh, L.J. Developmentally-regulated changes of Xist RNA levels in single preimplantation mouse embryos, as revealed by quantitative real-time PCR. Mol. Reprod. Dev. 2002, 61, 425–436. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liyanage, V.R.B.; Olson, C.O.; Zachariah, R.M.; Davie, J.R.; Rastegar, M. DNA Methylation Contributes to the Differential Expression Levels of Mecp2 in Male Mice Neurons and Astrocytes. Int. J. Mol. Sci. 2019, 20, 1845. https://doi.org/10.3390/ijms20081845
Liyanage VRB, Olson CO, Zachariah RM, Davie JR, Rastegar M. DNA Methylation Contributes to the Differential Expression Levels of Mecp2 in Male Mice Neurons and Astrocytes. International Journal of Molecular Sciences. 2019; 20(8):1845. https://doi.org/10.3390/ijms20081845
Chicago/Turabian StyleLiyanage, Vichithra R.B., Carl O. Olson, Robby M. Zachariah, James R. Davie, and Mojgan Rastegar. 2019. "DNA Methylation Contributes to the Differential Expression Levels of Mecp2 in Male Mice Neurons and Astrocytes" International Journal of Molecular Sciences 20, no. 8: 1845. https://doi.org/10.3390/ijms20081845
APA StyleLiyanage, V. R. B., Olson, C. O., Zachariah, R. M., Davie, J. R., & Rastegar, M. (2019). DNA Methylation Contributes to the Differential Expression Levels of Mecp2 in Male Mice Neurons and Astrocytes. International Journal of Molecular Sciences, 20(8), 1845. https://doi.org/10.3390/ijms20081845