ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Identification and Sequence Analysis of ZmASR Proteins
2.2. Expression of the ZmASR Genes in Different Tissues
2.3. Sub-Cellular Localization and Interaction Analysis of ZmASRs
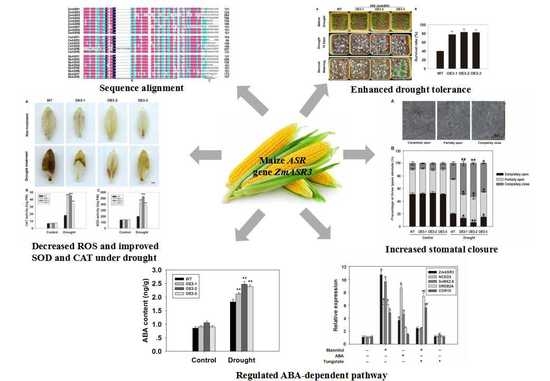
2.4. Characterization of the Function of ZmASRs in Drought Tolerance
2.5. Overexpression of ZmASR3 Enhances Drought Tolerance in Arabidopsis
2.6. Overexpression of ZmASR3 Increases Stomatal Closure
2.7. Overexpression of ZmASR3 Increases Drought Tolerance by Decreasing H2O2 Accumulation and Increasing Superoxide Dismutase (SOD) and Catalase (CAT) Activities
2.8. ZmASR3 Regulates the Expression of ROS-Related and Stress-Responsive Genes under Drought Conditions
2.9. Overexpression of ZmASR3 Increased Endogenous ABA Levels and Reduced Sensitivity to Exogenous ABA
2.10. Overexpression of ZmASR3 Enhances Drought Tolerance via an ABA-Dependent Pathway
3. Discussion
4. Methods and Materials
4.1. Identification and Multiple Sequence Alignment of ASR Proteins
4.2. Plant Growing Conditions and Stress Treatments
4.3. Constructs and Plant Transformation
4.4. Sub-Cellular Localization Assay
4.5. Yeast Two-Hybrid Assays
4.6. Total RNA Isolation and qRT-PCR analysis
4.7. Sensitivity Analysis of Exogenous ABA
4.8. DAB Staining, Determination of Physiological and Biochemical Activities, and ABA Content
4.9. Measurement of RWC and Proline and MDA Contents
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Du, Y.; Liu, L.; Li, M.; Fang, S.; Shen, X.; Chu, J.; Zhang, Z. UNBRANCHED3 regulates branching by modulating cytokinin biosynthesis and signaling in maize and rice. New Phytol. 2017, 214, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Shengxue, L.; Xianglan, W.; Hongwei, W.; Haibo, X.; Xiaohong, Y.; Jianbing, Y.; Jiansheng, L.; Lam-Son Phan, T.; Kazuo, S.; Kazuko, Y.S. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013, 9, e1003790. [Google Scholar]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J.; Muday, G.K. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA–mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wei, Z.; Gong, X.; Cheng, B. Expression of ZmHDZ4, a Maize Homeodomain-Leucine Zipper I Gene, Confers Tolerance to Drought Stress in Transgenic Rice. Plant Mol. Biol. Report. 2016, 34, 845–853. [Google Scholar] [CrossRef]
- Hong, E.; Lim, C.W.; Han, S.W.; Lee, S.C. Functional Analysis of the Pepper Ethylene-Responsive Transcription Factor, CaAIEF1, in Enhanced ABA Sensitivity and Drought Tolerance. Front. Plant Sci. 2017, 8, 1407. [Google Scholar] [CrossRef]
- Bao-Cai, T.; Joseph, L.M.; Wen-Tao, D.; Lijuan, L.; Qin-Bao, L.; Kenneth, C.; Mccarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. Cell Mol. Biol. 2010, 35, 44–56. [Google Scholar]
- Ic, K.J.M.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jdg, J.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. Embo. J. 2014, 22, 2623–2633. [Google Scholar]
- Iusem, N.D.; Bartholomew, D.M.; Hitz, W.D.; Scolnik, P.A. Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiol. 1993, 102, 1353–1354. [Google Scholar] [CrossRef]
- Dominguez, P.G.; Carrari, F. ASR1 transcription factor and its role in metabolism. Plant Signal. Behav. 2015, 10, e992751. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.; Lee, Y.H.; Kim, Y.-K.; Nahm, B.H; Song, S.I. Abiotic stress responsive rice ASR1 and ASR3 exhibit different;tissue-dependent sugar and hormone-sensitivities. Mol. Cells 2013, 35, 421–435. [Google Scholar] [CrossRef]
- Frankel, N.; Carrari, F.; Hasson, E.; Iusem, N.D. Evolutionary history of the Asr gene family. Gene 2006, 378, 74–83. [Google Scholar] [CrossRef]
- Laetitia, V.; Marie-Pierre, J.; Denise, G.; Hélène, C.; Sophie, B.; Françoise, G.; Benoît, V.; Jacques, T.; Guillaume, T.; Matthieu, F. The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol. 2011, 157, 917–936. [Google Scholar]
- Wei, H.; Chao, H.; Xiaomin, D.; Shiyi, Z.; Lihong, C.; Yin, L.; Cheng, W.; Zhanbing, M.; Qianqian, Y.; Yan, W. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 2013, 36, 1449–1464. [Google Scholar]
- Wong, C.E.; Yong, L.; Aurelie, L.; David, G.; Paulo, N.; Brett, W.; Claudia, D.; G Brian, G.; Gray, G.R.; Weretilnyk, E.A. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006, 140, 1437–1450. [Google Scholar] [CrossRef]
- Laura, M.; Nicolás, F.; Gustavo, G.; Demergasso, M.J.; Pietrasanta, L.I.; Iusem, N.D. Dimerization and DNA-binding of ASR1, a small hydrophilic protein abundant in plant tissues suffering from water loss. Biochem. Biophys. Res. Commun. 2007, 352, 831–835. [Google Scholar]
- Kim, I.S.; Kim, Y.S.; Yoon, H.S. Rice ASR1 protein with reactive oxygen species scavenging and chaperone-like activities enhances acquired tolerance to abiotic stresses in Saccharomyces cerevisiae. Mol. Cells 2012, 33, 285–293. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, H.; Su, H.; Xia, K.; Jian, S.; Zhang, M. Ipomoea pes-caprae IpASR improves salinity and drought tolerance in transgenic Escherichia coli and Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2252. [Google Scholar] [CrossRef]
- Jian-Ye, C.; Du-Juan, L.; Yue-Ming, J.; Ming-Lei, Z.; Wei, S.; Jian-Fei, K.; Wang-Jin, L. Molecular characterization of a strawberry FaASR gene in relation to fruit ripening. PLoS ONE 2011, 6, e24649. [Google Scholar]
- Vivekanand, T.; Amit Kumar, C.; Avinash, M.; Bhavanath, J. Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiate enhances salinity and drought endurance in transgenic groundnut (Arachis hypogaea) and acts as a transcription factor. PLoS ONE 2015, 10, e0131567. [Google Scholar]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Zvia, K.; Dudy, B.Z. Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta 2008, 227, 1213–1219. [Google Scholar]
- Murata, Y.; Mori, I.C.; Munemasa, S. Diverse Stomatal Signaling and the Signal Integration Mechanism. Annu. Rev. Plant Biol. 2015, 66, 369. [Google Scholar] [CrossRef]
- Aying, Z.; Jun, Z.; Jianhua, Z.; Nenghui, Y.; Hong, Z.; Mingpu, T.; Mingyi, J. Nitric oxide mediates brassinosteroid-induced ABA biosynthesis involved in oxidative stress tolerance in maize leaves. Plant Cell Physiol. 2011, 52, 181. [Google Scholar]
- June-Sik, K.; Junya, M.; Satoshi, K.; Kyonoshin, M.; Jun, N.; Kazuo, N.; Nobutaka, M.; Yuko, T.; Masaru, O.T.; Youichi, K. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 2012, 24, 3393–3405. [Google Scholar]
- Liang, J.; Guo, S.; Sun, B.; Liu, Q.; Chen, X.; Peng, H.; Zhang, Z.; Xie, Q. Constitutive expression of REL1 confers the rice response to drought stress and abscisic acid. Rice 2018. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, X.; Jing, R.; Chang, X.; Xie, H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot. 2011, 62, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, H.; Zhang, B.; Bai, X.; Zhou, C. Physiological and Molecular Changes of Detached Wheat Leaves in Responding to Various Treatments. J. Integr. Plant Biol. 2012, 54, 10. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, X.; Yu, Z. Transformation of Salt-tolerance Gene DREB2A into Maize Mediated by Agrobacterium Tumefaciens. Mol. Plant Breed. 2013, 11, 48–52. [Google Scholar]
- Anja, T.; Hincha, D.K. A mechanistic model of COR15 protein function in plant freezing tolerance: Integration of structural and functional characteristics. Plant Signal. Behav. 2014, 9, e977722. [Google Scholar]
- Yossi, K.; Ayelet, G.; Zvia, K.; Michele, Z.; Scolnik, P.A.; Dudy, B.Z. The water- and salt-stress-regulated Asr1 (abscisic acid stress ripening) gene encodes a zinc-dependent DNA-binding protein. Biochem. J. 2004, 381, 373. [Google Scholar]
- Birsen, C.; Alice, A.; Cécile, G.; Amélie, S.; Serge, D.; Rossitza, A. A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 2003, 15, 2165–2180. [Google Scholar]
- Padmanabhan, V.; Dias, D.M.A.L.; Newton, R.J. Expression analysis of a gene family in loblolly pine (Pinus taeda L.) induced by water deficit stress. Plant Mol. Biol. 1997, 35, 801–807. [Google Scholar] [CrossRef]
- Jorge, P.D.; Tsung-Meng, W.; Ricardo, P.D.; Simón, R.L.; Chwan-Yang, H.; Casaretto, J.A. Organ- and stress-specific expression of the ASR genes in rice. Plant Cell Rep. 2014, 33, 61–73. [Google Scholar]
- Li, X.; Hou, S.; Gao, Q.; Zhao, P.; Chen, S.; Qi, D.; Lee, B.H.; Cheng, L.; Liu, G. LcSAIN1, a Novel Salt-Induced Gene from SheepGrass, Confers Salt Stress Tolerance in Transgenic Arabidopsis and Rice. Plant Cell Physiol. 2013, 54, 1172–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zsigmond, L.; Szepesi, Á.; Tari, I.; Rigó, G.; Király, A.; Szabados, L. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Sci. 2012, 182, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Jing, N.; Xianghua, L.; Hicks, L.M.; Lizhong, X. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010, 152, 876–890. [Google Scholar]
- Aoyagi, L.N.; Lopes-Caitar, V.S.; Carvalho, M.C.C.G.D.; Darben, L.M.; Polizel-Podanosqui, A.; Kuwahara, M.K.; Nepomuceno, A.L.; Abdelnoor, R.V. Genomic and transcriptomic characterization of the transcription factor family R2R3-MYB in soybean and its involvement in the resistance responses to Phakopsora pachyrhizi. Plant Sci. Int. J. Exp. Plant Biol. 2014, 229, 32–42. [Google Scholar] [CrossRef]
- Polle, A. Dissecting the superoxide dismutase-ascorbate-glutathione-pathway in chloroplasts by metabolic modeling. Computer simulations as a step towards flux analysis. Plant Physiol. 2001, 126, 445–462. [Google Scholar] [CrossRef]
- Ron, M.; Sandy, V.; Martin, G.; Frank, V.B. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar]
- Diaz-Vivancos, P.; Faize, M.; Barba-Espin, G.; Faize, L.; Petri, C.; Hernández, J.A.; Burgos, L. Ectopic expression of cytosolic superoxide dismutase and ascorbate peroxidase leads to salt stress tolerance in transgenic plums. Plant Biotechnol. J. 2013, 11, 976–985. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K.H.M. Drought Stress in Plants: An Overview. In Plant Responses to Drought Stress; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Gang, L.; Yu, D. Activated Expression of WRKY57 Confers Drought Tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Shinozaki, K.; Yamaguchishinozaki, K. Plant gene networks in osmotic stress response: From genes to regulatory networks. Methods Enzymol. 2007, 428, 109–128. [Google Scholar] [PubMed]
- Meyer, K.; Leube, M.P.; Grill, E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 1994, 264, 1452–1455. [Google Scholar] [CrossRef]
- Endo, A.; Egawa, C.; Oohashi, M.; Meguro-Maoka, A.; Shimosaka, E.; Sato, Y. Ectopic expression of mutated type 2C protein phosphataseOsABI-LIKE2decreases abscisic acid sensitivity in Arabidopsis and rice. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, Q.; Long, C.; Liang, Y.; Wu, J. Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 2018, 40, 9. [Google Scholar] [CrossRef]
- Lim, C.W.; Lee, S.C. Pepper protein phosphatase type 2C, CaADIP1 and its interacting partner CaRLP1 antagonistically regulate ABA signalling and drought response. Plant Cell Environ. 2016, 39, 1559–1575. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 764–775. [Google Scholar]
- Polle, A.; Otter, T.; Seifert, F. Apoplastic Peroxidases and Lignification in Needles of Norway Spruce (Picea abies L.). Plant Physiol. 1994, 106, 53–60. [Google Scholar] [CrossRef]
- Jiu-Feng, L.; Jun, D.; Bi-Feng, Y.; Yu-Qi, F. Magnetic solid phase extraction coupled with in situ derivatization for the highly sensitive determination of acidic phytohormones in rice leaves by UPLC-MS/MS. Analyst 2014, 139, 5605. [Google Scholar]
- Lu, X.; Yang, L.; Yu, M.; Lai, J.; Wang, C.; Mcneil, D.; Zhou, M.; Yang, C. A novel Zea mays ssp. mexicana L. MYC-type ICE -like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 113, 78–88. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Yang, Z.; Qing, M.; Xiaolei, J.; Xiaojian, P.; Jinyang, L.; Lin, D.; Hanwei, Y.; Lei, S.; Haiyang, J.; Beijiu, C. A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 2014, 55, 1142–1156. [Google Scholar]
- Zhang, S.; Li, N.; Gao, F.; Yang, A.; Zhang, J. Over-expression of TsCBF1 gene confers improved drought tolerance in transgenic maize. Mol. Breed. 2010, 26, 455–465. [Google Scholar] [CrossRef]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Jiang, Y.; Du, M.; Li, B.; Chen, L.; Chen, M.; Jin, D.; Wu, J. ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2278. https://doi.org/10.3390/ijms20092278
Liang Y, Jiang Y, Du M, Li B, Chen L, Chen M, Jin D, Wu J. ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2019; 20(9):2278. https://doi.org/10.3390/ijms20092278
Chicago/Turabian StyleLiang, Yani, Yingli Jiang, Ming Du, Baoyan Li, Long Chen, Mingchao Chen, Demiao Jin, and Jiandong Wu. 2019. "ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis" International Journal of Molecular Sciences 20, no. 9: 2278. https://doi.org/10.3390/ijms20092278
APA StyleLiang, Y., Jiang, Y., Du, M., Li, B., Chen, L., Chen, M., Jin, D., & Wu, J. (2019). ZmASR3 from the Maize ASR Gene Family Positively Regulates Drought Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences, 20(9), 2278. https://doi.org/10.3390/ijms20092278