Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms
Abstract
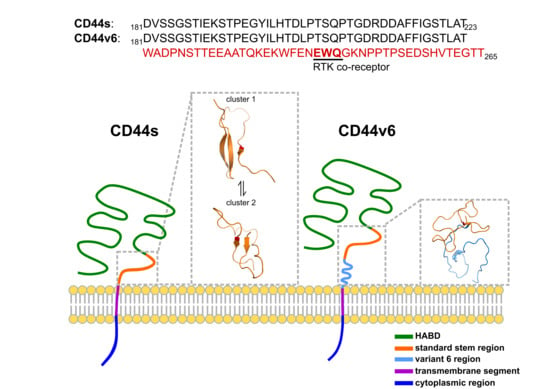
:1. Introduction
2. Results
2.1. CD44s Stem Region Adopted Predominantly Two Conformations, Whereas CD44v6 Mainly Adopted One
2.2. The Standard Stem Region of CD44v6 Was Much More Flexible than CD44s
2.3. Phe215 Displayed a High Tendency to Form β-Strands in CD44s and CD44v6, Whereas the CD44v6 Stem Region Exhibited Fewer β-Sheets than CD44s Did
2.4. CD44s and CD44v6 Utilized Phe125 with Different Interaction Partners to Form Hydrogen Bonds (Hbonds) to Stabilize β-Sheets
2.5. Phe215 in CD44v6 Interacted with Surrounding Hydrophobic Residues More than CD44s, Leading to Becoming Blocked
3. Materials and Methods
3.1. Protein Model Preparation
3.2. Molecular Dynamics Simulations
3.3. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HA | Hyaluronan |
| HABD | HA-binding domain |
| RTKs | Receptor tyrosine kinases |
| VEGFR-2 | vascular endothelial growth factor receptor 2 |
| NMR | Nuclear magnetic resonance |
| PDB | Protein Data Bank |
| MD | Molecular dynamics |
| RMSD | Root mean square deviation |
| Rg | Radius of gyration |
| RMSF | Root mean square fluctuation |
| SASA | Solvent-accessible surface area |
References
- Stamenkovic, I.; Amiot, M.; Pesando, J.M.; Seed, B. A lymphocyte molecule implicated in lymph node homing is a member of the cartilage link protein family. Cell 1989, 56, 1057–1062. [Google Scholar] [CrossRef]
- Aruffo, A.; Stamenkovic, I.; Melnick, M.; Underhill, C.B.; Seed, B. CD44 is the principal cell surface receptor for hyaluronate. Cell 1990, 61, 1303–1313. [Google Scholar] [CrossRef]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Dzwonek, J.; Wilczynski, G.M. CD44: Molecular interactions, signaling and functions in the nervous system. Front. Cell. Neurosci. 2015, 9, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerji, S.; Day, A.J.; Kahmann, J.D.; Jackson, D.G. Characterization of a Functional Hyaluronan-Binding Domain from the Human CD44 Molecule Expressed inEscherichia coli. Protein Expr. Purif. 1998, 14, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Ford, R.; Thomas, P.; Nachman, A.; Steele, G., Jr.; Jessup, J. CD44 participates in the adhesion of human colorectal carcinoma cells to laminin and type IV collagen. Surg. Oncol. 1993, 2, 255–264. [Google Scholar] [CrossRef]
- Jalkanen, S.; Jalkanen, M. Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J. Cell Biol. 1992, 116, 817–825. [Google Scholar] [CrossRef]
- Screaton, G.R.; Bell, M.V.; Jackson, D.G.; Cornelis, F.B.; Gerth, U.; Bell, J. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc. Natl. Acad. Sci. USA 1992, 89, 12160–12164. [Google Scholar] [CrossRef] [Green Version]
- Naor, D.; Wallach-Dayan, S.B.; Zahalka, M.A.; Sionov, R.V. Involvement of CD44, a molecule with a thousand faces, in cancer dissemination. In Hyaluronan in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 127–146. [Google Scholar]
- Acharya, P.S.; Majumdar, S.; Jacob, M.; Hayden, J.; Mrass, P.; Weninger, W.; Assoian, R.K.; Puré, E. Fibroblast migration is mediated by CD44-dependent TGFβ activation. J. Cell Sci. 2008, 121, 1393–1402. [Google Scholar] [CrossRef] [Green Version]
- Stamenkovic, I.; Aruffo, A.; Amiot, M.; Seed, B. The hematopoietic and epithelial forms of CD44 are distinct polypeptides with different adhesion potentials for hyaluronate-bearing cells. EMBO J. 1991, 10, 343–348. [Google Scholar] [CrossRef]
- Bartolazzi, A.; Nocks, A.; Aruffo, A.; Spring, F.; Stamenkovic, I. Glycosylation of CD44 is implicated in CD44-mediated cell adhesion to hyaluronan. J. Cell Biol. 1996, 132, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Modrell, B.; Greenfield, B.; Bartolazzi, A.; Stamenkovic, I.; Peach, R.; Jackson, D.G.; Spring, F.; Aruffo, A. Regulation of CD44 binding to hyaluronan by glycosylation of variably spliced exons. J. Cell Biol. 1995, 131, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.L.; Jackson, D.G.; Simon, J.C.; Tanczos, E.; Peach, R.; Modrell, B.; Stamenkovic, I.; Plowman, G.; Aruffo, A. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J. Cell Biol. 1995, 128, 687–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, B.; Wang, W.-C.; Marquardt, H.; Piepkorn, M.; Wolff, E.A.; Aruffo, A.; Bennett, K.L. Characterization of the heparan sulfate and chondroitin sulfate assembly sites in CD44. J. Biol. Chem. 1999, 274, 2511–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, P.; Schwärzler, C.; Günthert, U. CD44 isoforms during differentiation and development. Bioessays 1995, 17, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 cell adhesion molecules. Mol. Pathol. 1999, 52, 189. [Google Scholar] [CrossRef] [Green Version]
- Günthert, U.; Hofmann, M.; Rudy, W.; Reber, S.; Zöller, M.; Hauβmann, I.; Matzku, S.; Wenzel, A.; Ponta, H.; Herrlich, P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 1991, 65, 13–24. [Google Scholar] [CrossRef]
- Hofmann, M.; Rudy, W.; Zöller, M.; Tölg, C.; Ponta, H.; Herrlich, P.; Günthert, U. CD44 splice variants confer metastatic behavior in rats: Homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991, 51, 5292–5297. [Google Scholar]
- Pries, R.; Wittkopf, N.; Trenkle, T.; Nitsch, S.M.; Wollenberg, B. Potential stem cell marker CD44 is constitutively expressed in permanent cell lines of head and neck cancer. In Vivo 2008, 22, 89–92. [Google Scholar]
- Zöller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.J.; Dosch, J.; Simeone, D.M. Pancreatic cancer stem cells. J. Clin. Oncol. 2008, 26, 2806–2812. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Balch, C.; Chan, M.W.; Lai, H.-C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.; Nephew, K.P. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeilstra, J.; Joosten, S.; Van Andel, H.; Tolg, C.; Berns, A.; Snoek, M.; Van De Wetering, M.; Spaargaren, M.; Clevers, H.; Pals, S. Stem cell CD44v isoforms promote intestinal cancer formation in Apc (min) mice downstream of Wnt signaling. Oncogene 2014, 33, 665. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Zuo, X.; Wei, D. Concise review: Emerging role of CD44 in cancer stem cells: A promising biomarker and therapeutic target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis. 2019, 10, 30. [Google Scholar] [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Leuci, V.; Casucci, M.; Grignani, G.; Rotolo, R.; Rossotti, U.; Vigna, E.; Gammaitoni, L.; Mesiano, G.; Fiorino, E.; Donini, C. CD44v6 as innovative sarcoma target for CAR-redirected CIK cells. Oncoimmunology 2018, 7, e1423167. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, P.J.; Sousa, F.; Ferreira, D.; Pereira, C.; Nestor, M.; Oliveira, C.; Granja, P.L.; Sarmento, B. Fab-conjugated PLGA nanoparticles effectively target cancer cells expressing human CD44v6. Acta Biomater. 2018, 81, 208–218. [Google Scholar] [CrossRef]
- Odenthal, J.; Rijpkema, M.; Bos, D.; Wagena, E.; Croes, H.; Grenman, R.; Boerman, O.; Takes, R.; Friedl, P. Targeting CD44v6 for fluorescence-guided surgery in head and neck squamous cell carcinoma. Sci. Rep. 2018, 8, 10467. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V. CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer 2010, 46, 1271–1277. [Google Scholar] [CrossRef] [PubMed]
- Orian-Rousseau, V.; Morrison, H.; Matzke, A.; Kastilan, T.; Pace, G.; Herrlich, P.; Ponta, H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol. Biol. Cell 2007, 18, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Tremmel, M.; Matzke, A.; Albrecht, I.; Laib, A.M.; Olaku, V.; Ballmer-Hofer, K.; Christofori, G.; Héroult, M.; Augustin, H.G.; Ponta, H. A CD44v6 peptide reveals a role of CD44 in VEGFR-2 signaling and angiogenesis. Blood 2009, 114, 5236–5244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, A.; Herrlich, P.; Ponta, H.; Orian-Rousseau, V. A five-amino-acid peptide blocks Met-and Ron-dependent cell migration. Cancer Res. 2005, 65, 6105–6110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orian-Rousseau, V. CD44 acts as a signaling platform controlling tumor progression and metastasis. Front. Immunol. 2015, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Vuorio, J.; Vattulainen, I.; Martinez-Seara, H. Atomistic fingerprint of hyaluronan–CD44 binding. PLoS Comput. Biol. 2017, 13, e1005663. [Google Scholar] [CrossRef] [Green Version]
- Guvench, O. Revealing the mechanisms of protein disorder and N-glycosylation in CD44-hyaluronan binding using molecular simulation. Front. Immunol. 2015, 6, 305. [Google Scholar] [CrossRef] [Green Version]
- Favreau, A.J.; Faller, C.E.; Guvench, O. CD44 receptor unfolding enhances binding by freeing basic amino acids to contact carbohydrate ligand. Biophys. J. 2013, 105, 1217–1226. [Google Scholar] [CrossRef] [Green Version]
- Jamison, F.W., II; Foster, T.J.; Barker, J.A.; Hills, R.D., Jr.; Guvench, O. Mechanism of binding site conformational switching in the CD44–hyaluronan protein–carbohydrate binding interaction. J. Mol. Biol. 2011, 406, 631–647. [Google Scholar] [CrossRef]
- Patel, S.; Shaikh, F.; Devaraji, V.; Radadiya, A.; Shah, K.; Shah, A.; Rawal, R. Insights into the structural perturbations of spliced variants of CD44: A modeling and simulation approach. J. Biomol. Struct. Dyn. 2017, 35, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Teriete, P.; Banerji, S.; Noble, M.; Blundell, C.D.; Wright, A.J.; Pickford, A.R.; Lowe, E.; Mahoney, D.J.; Tammi, M.I.; Kahmann, J.D. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol. Cell 2004, 13, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the Cd44–hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Ogino, S.; Umemoto, R.; Sakakura, M.; Kajiwara, M.; Sugahara, K.N.; Hayasaka, H.; Miyasaka, M.; Terasawa, H.; Shimada, I. Ligand-induced structural changes of the CD44 hyaluronan-binding domain revealed by NMR. J. Biol. Chem. 2006, 281, 40089–40095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.-K.; Finzel, B. High-resolution crystal structures of alternate forms of the human CD44 hyaluronan-binding domain reveal a site for protein interaction. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1155–1161. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, S.A.; Dror, R.O. Molecular dynamics simulation for all. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef] [Green Version]
- Śledź, P.; Caflisch, A. Protein structure-based drug design: From docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018, 48, 93–102. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Rauscher, S.; Pomès, R. Molecular simulations of protein disorder. Biochem. Cell Biol. 2010, 88, 269–290. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Mol. Biol. 2002, 9, 646. [Google Scholar] [CrossRef]
- Matzke-Ogi, A.; Jannasch, K.; Shatirishvili, M.; Fuchs, B.; Chiblak, S.; Morton, J.; Tawk, B.; Lindner, T.; Sansom, O.; Alves, F.; et al. Inhibition of Tumor Growth and Metastasis in Pancreatic Cancer Models by Interference with CD44v6 Signaling. Gastroenterology 2016, 150, 513–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takazoe, K.; Foti, R.; Tesch, G.H.; Hurst, L.A.; Lan, H.Y.; Atkins, R.C.; Nikolic-Paterson, D.J. Up-regulation of the tumour-associated marker CD44V6 in experimental kidney disease. Clin. Exp. Immunol. 2000, 121, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Hallouin, F.; Goupille, C.; leCabellec, M.; Bara, J.; lePendu, J. Expression of A and H blood-group and of CD44 antigens during chemical rat colonic carcinogenesis. Glycoconj. J. 1997, 14, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Khor, B.Y.; Tye, G.J.; Lim, T.S.; Choong, Y.S. General overview on structure prediction of twilight-zone proteins. Theor. Biol. Med. Model. 2015, 12, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padmanabhan, S. Handbook of Pharmacogenomics and Stratified Medicines/Edited by Sandosh Padmanabhan, Institute of Cardiovascular and Medical Sciences; University of Glasgow: Glasgow, UK, 2014. [Google Scholar]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perczel, A.; Gaspari, Z.; Csizmadia, I.G. Structure and stability of beta-pleated sheets. J. Comput. Chem. 2005, 26, 1155–1168. [Google Scholar] [CrossRef]
- Zhao, Y.L.; Wu, Y.D. A theoretical study of beta-sheet models: Is the formation of hydrogen-bond networks cooperative? J. Am. Chem. Soc. 2002, 124, 1570–1571. [Google Scholar] [CrossRef]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Ranganathan, S.; Nakai, K.; Schonbach, C. Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Minor, D.L.; Kim, P.S. Measurement of the Beta-Sheet-Forming Propensities of Amino-Acids. Nature 1994, 367, 660–663. [Google Scholar] [CrossRef]
- Street, A.G.; Mayo, S.L. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc. Natl. Acad. Sci. USA 1999, 96, 9074–9076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen bonds in proteins: Role and strength. eLS 2010. [Google Scholar] [CrossRef]
- Ho, B.K.; Coutsias, E.A.; Seok, C.; Dill, K.A. The flexibility in the proline ring couples to the protein backbone. Protein Sci. 2005, 14, 1011–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burley, S.; Petsko, G.A. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Tolg, C.; Hofmann, M.; Herrlich, P.; Ponta, H. Splicing Choice from 10 Variant Exons Establishes Cd44 Variability. Nucleic Acids Res. 1993, 21, 1225–1229. [Google Scholar] [CrossRef] [Green Version]
- Screaton, G.R.; Bell, M.V.; Bell, J.I.; Jackson, D.G. The Identification of a New Alternative Exon with Highly Restricted Tissue Expression in Transcripts Encoding the Mouse Pgp-1 (Cd44) Homing Receptor - Comparison of All 10 Variable Exons between Mouse, Human, and Rat. J. Biol. Chem. 1993, 268, 12235–12238. [Google Scholar]
- Yang, J.Y.; Yan, R.X.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.Y.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A Method to Identify Protein Sequences That Fold into a Known 3-Dimensional Structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with 3-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone phi, psi and Side-Chain chi(1) and chi(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Feig, M.; Brooks, C.L. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004, 126, 698–699. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald—an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]








| van der Waals Energy | Electrostatic Energy | Interaction Energy | |
|---|---|---|---|
| CD44s, cluster 1 | −1.196 ± 1.058 | −0.1625 ± 0.3979 | −1.358 ± 1.243 |
| CD44s, cluster 2 | −1.534 ± 0.8957 | 0.1097 ± 0.2475 | −1.424 ± 0.9442 |
| CD44v6, cluster 1 | −2.394 ± 1.445 | −0.08138 ± 0.2759 | −2.476 ± 1.485 |
| SASA (Å2) | Buried Percentage (%) | |
|---|---|---|
| CD44s, cluster 1 | 114.30 ± 31.24 | 39.85 ± 16.44% |
| CD44s, cluster 2 | 92.19 ± 31.64 | 51.48 ± 16.65% |
| CD44v6, cluster 1 | 55.18 ± 36.08 | 70.96 ± 18.99% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.-L.; Li, D.; Lu, T.-X.; Chang, S.-W. Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms. Int. J. Mol. Sci. 2020, 21, 336. https://doi.org/10.3390/ijms21010336
Chen K-L, Li D, Lu T-X, Chang S-W. Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms. International Journal of Molecular Sciences. 2020; 21(1):336. https://doi.org/10.3390/ijms21010336
Chicago/Turabian StyleChen, Kun-Lin, Deng Li, Ting-Xuan Lu, and Shu-Wei Chang. 2020. "Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms" International Journal of Molecular Sciences 21, no. 1: 336. https://doi.org/10.3390/ijms21010336
APA StyleChen, K. -L., Li, D., Lu, T. -X., & Chang, S. -W. (2020). Structural Characterization of the CD44 Stem Region for Standard and Cancer-Associated Isoforms. International Journal of Molecular Sciences, 21(1), 336. https://doi.org/10.3390/ijms21010336