CaCML13 Acts Positively in Pepper Immunity Against Ralstonia solanacearum Infection Forming Feedback Loop with CabZIP63
Abstract
:1. Introduction
2. Results
2.1. The CML Gene Was Upregulated by R. solanacearum Inoculation
2.2. The CML Is CaCML13 Exhibiting High Sequence Similarities to CMLs in Other Plant Species
2.3. CaCML13 Is a Protein Distributes in the Whole Cell Including Plasma Membrane, Cytoplasm and the Nucleus
2.4. The Silencing of CaCML13 Significantly Enhanced Susceptibility to RSI
2.5. Transient Overexpression of CaCML13 Triggered Intensive Hypersensitivity Reaction (HR)-Mimicked Cell Death
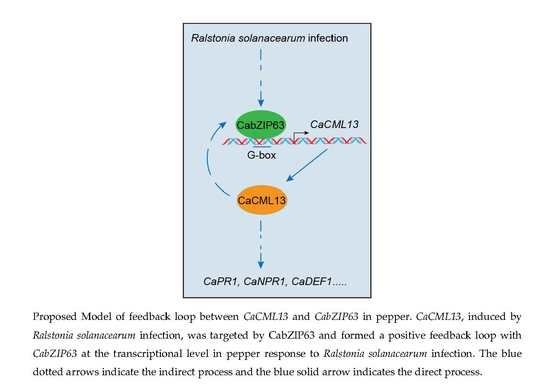
2.6. The G-Box-Containing CaCML13 Promoter Are Directly Bound by CabZIP63
2.7. The Interrelationship between CabZIP63 and CaCML13 at the Transcriptional Level
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. The Vectors Construction
4.3. VIGS Assay
4.4. Transient Overexpression of CaCML13-HA in Pepper Leaves
4.5. R. solanacearum Growth and RSI
4.6. Subcellular Localization
4.7. ChIP Analysis
4.8. qRT-PCR Assay
4.9. Measurement of Ion Conductivity
4.10. Histochemical Staining Assay
4.11. Immunoblot Analysis
4.12. Calculation of Disease Index of Bacterial Wilt
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, J.; Meng, X. Phosphoregulation of Ca2+ influx in plant immunity. Trends Plant Sci. 2019, 24, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, B.W.; Du, L. Calcium signaling: Decoding mechanism of calcium signatures. New Phytol. 2018, 217, 1394–1396. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 2014, 9, e973818. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, K.; Kudla, J. Calcium decoding mechanisms in plants. Biochimie 2011, 93, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Li, X.; Fan, T.; Luo, H.; Xie, C.; Zhou, Y.; Tian, C.E. The calmodulin-binding protein IQM1 interacts with CATALASE2 to affect pathogen defense. Plant Physiol. 2019, 181, 1314–1327. [Google Scholar] [CrossRef]
- Campe, R.; Langenbach, C.; Leissing, F.; Popescu, G.V.; Popescu, S.C.; Goellner, K.; Beckers, G.J.; Conrath, U. ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol. 2016, 209, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Batistic, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [Green Version]
- Wan, D.; Li, R.; Zou, B.; Zhang, X.; Cong, J.; Wang, R.; Xia, Y.; Li, G. Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 2012, 31, 1269–1281. [Google Scholar] [CrossRef]
- Sun, T.; Huang, J.; Xu, Y.; Verma, V.; Jing, B.; Sun, Y.; Ruiz Orduna, A.; Tian, H.; Huang, X.; Xia, S.; et al. Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol. Plant 2020, 13, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Naveed, Z.A.; Bibi, S.; Ali, G.S. The phytophthora RXLR effector Avrblb2 modulates plant immunity by interfering with Ca2+ signaling pathway. Front. Plant Sci. 2019, 10, 374. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wagener, N.; McLellan, H.; Boevink, P.C.; Hua, C.; Birch, P.R.J.; Brunner, F. Phytophthora infestans RXLR effector SFI5 requires association with calmodulin for PTI/MTI suppressing activity. New Phytol. 2018, 219, 1433–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boonburapong, B.; Buaboocha, T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Meng, D.; Zhang, J.; Cheng, L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malusx domestica). Plant Physiol. Biochem. 2019, 139, 600–612. [Google Scholar] [CrossRef]
- Leba, L.J.; Cheval, C.; Ortiz-Martin, I.; Ranty, B.; Beuzon, C.R.; Galaud, J.P.; Aldon, D. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 2012, 71, 976–989. [Google Scholar] [CrossRef]
- Ma, W.; Smigel, A.; Tsai, Y.C.; Braam, J.; Berkowitz, G.A. Innate immunity signaling: Cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008, 148, 818–828. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Perez, M.; Aldon, D.; Galaud, J.P. Respective contribution of CML8 and CML9, two arabidopsis calmodulin-like proteins, to plant stress responses. Plant Signal. Behav. 2017, 12, e1322246. [Google Scholar] [CrossRef]
- Zhu, X.; Robe, E.; Jomat, L.; Aldon, D.; Mazars, C.; Galaud, J.P. CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol. 2017, 58, 307–319. [Google Scholar]
- Chen, X.; Lu, L.; Qian, S.; Scalf, M.; Smith, L.M.; Zhong, X. Canonical and noncanonical actions of Arabidopsis histone deacetylases in ribosomal RNA processing. Plant Cell 2018, 30, 134–152. [Google Scholar] [CrossRef] [Green Version]
- Cheval, C.; Perez, M.; Leba, L.J.; Ranty, B.; Perochon, A.; Reichelt, M.; Mithofer, A.; Robe, E.; Mazars, C.; Galaud, J.P.; et al. PRR2, a pseudo-response regulator, promotes salicylic acid and camalexin accumulation during plant immunity. Sci. Rep. 2017, 7, 6979. [Google Scholar] [CrossRef] [Green Version]
- Hayward, A.C. Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu. Rev. Phytopathol. 1991, 29, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Ristaino, J.B.; Johnston, S.A. Ecologically based approaches to management of phytophthora blight on bell pepper. Plant Dis. 1999, 83, 1080–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, A.M.; Kousik, C.S.; Ritchie, D.F. Temperature sensitivity of the hypersensitive response of bell pepper to Xanthomonas axonopodis pv. vesicatoria. Phytopathology 2002, 92, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Liu, Z.; Yang, S.; Yang, T.; Liang, J.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 2016, 67, 2439–2451. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Yang, S.; Yang, T.; Liang, J.; Cheng, W.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci. Rep. 2016, 6, 22439. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Hwang, B.K. The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. 2012, 72, 843–855. [Google Scholar] [CrossRef]
- Kim, D.S.; Jeun, Y.; Hwang, B.K. The pepper patatin-like phospholipase CaPLP1 functions in plant cell death and defense signaling. Plant Mol. Biol. 2014, 84, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Lin, Y.M.; Chao, T.C.; Wang, J.F.; Liu, A.C.; Ho, F.I.; Cheng, C.P. Virus-induced gene silencing reveals the involvement of ethylene-, salicylic acid- and mitogen-activated protein kinase-related defense pathways in the resistance of tomato to bacterial wilt. Physiol. Plant. 2009, 136, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Lu, W.; Meng, F.; Wu, C.A.; Guo, X. Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J. Exp. Bot. 2012, 63, 3935–3951. [Google Scholar] [CrossRef] [PubMed]
- Mee Do, H.; Chul Lee, S.; Won Jung, H.; Hoon Sohn, K.; Kook Hwang, B. Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 2004, 166, 1297–1305. [Google Scholar] [CrossRef]
- Shen, H.; Cao, K.; Wang, X. AtbZIP16 and AtbZIP68, two new members of GBFs, can interact with other G group bZIPs in Arabidopsis thaliana. BMB Rep. 2008, 41, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Onate-Sanchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Droge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramonell, K.; Berrocal-Lobo, M.; Koh, S.; Wan, J.; Edwards, H.; Stacey, G.; Somerville, S. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 2005, 138, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [Green Version]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.H.; Hwang, B.K. Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol. 2015, 167, 307–322. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Choi, H.W.; Hwang, B.K. The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 2011, 156, 2011–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.; Doke, N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, H.M.; Hong, J.K.; Jung, H.W.; Kim, S.H.; Ham, J.H.; Hwang, B.K. Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Mol. Plant Microbe Interact. 2003, 16, 196–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Yang, S.; Yang, F.; Guan, D.; He, S. CaCBL1 acts as a positive regulator in pepper response to Ralstonia solanacearum. Mol. Plant Microbe Interact. 2020. [Google Scholar] [CrossRef]
- Liu, N.; Hake, K.; Wang, W.; Zhao, T.; Romeis, T.; Tang, D. CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1-mediated immunity. Plant Cell 2017, 29, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Schuck, S.; Wu, J.; Yang, P.; Doring, A.C.; Zeier, J.; Tsuda, K. A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 2018, 30, 2480–2494. [Google Scholar] [CrossRef] [Green Version]
- Casaretto, J.; Ho, T.H. The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 2003, 15, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Chung, H.J.; Thomas, T.L. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 1997, 11, 1237–1251. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Ohmiya, K.; Hattori, T. A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 1996, 9, 217–227. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, A.; Lei, Y.; Yang, S.; Wu, J.; Li, J.; Bao, B.; Cai, Y.; Wang, S.; Lin, J.; Wang, Y.; et al. CaC3H14 encoding a tandem CCCH zinc finger protei Iis directly targeted by CaWRKY40 and positively regulates the response of pepper to inoculation by Ralstonia solanacearum. Mol. Plant Pathol. 2018, 19, 2221–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hwang, I.S.; Hwang, B.K. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 2011, 155, 447–463. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.W.; Lee, B.G.; Kim, N.H.; Park, Y.; Lim, C.W.; Song, H.K.; Hwang, B.K. A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol. 2008, 148, 383–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, L.; Yang, S.; Guan, D.; He, S. CaCML13 Acts Positively in Pepper Immunity Against Ralstonia solanacearum Infection Forming Feedback Loop with CabZIP63. Int. J. Mol. Sci. 2020, 21, 4186. https://doi.org/10.3390/ijms21114186
Shen L, Yang S, Guan D, He S. CaCML13 Acts Positively in Pepper Immunity Against Ralstonia solanacearum Infection Forming Feedback Loop with CabZIP63. International Journal of Molecular Sciences. 2020; 21(11):4186. https://doi.org/10.3390/ijms21114186
Chicago/Turabian StyleShen, Lei, Sheng Yang, Deyi Guan, and Shuilin He. 2020. "CaCML13 Acts Positively in Pepper Immunity Against Ralstonia solanacearum Infection Forming Feedback Loop with CabZIP63" International Journal of Molecular Sciences 21, no. 11: 4186. https://doi.org/10.3390/ijms21114186
APA StyleShen, L., Yang, S., Guan, D., & He, S. (2020). CaCML13 Acts Positively in Pepper Immunity Against Ralstonia solanacearum Infection Forming Feedback Loop with CabZIP63. International Journal of Molecular Sciences, 21(11), 4186. https://doi.org/10.3390/ijms21114186