Black Soldier Fly Larvae Adapt to Different Food Substrates through Morphological and Functional Responses of the Midgut
Abstract
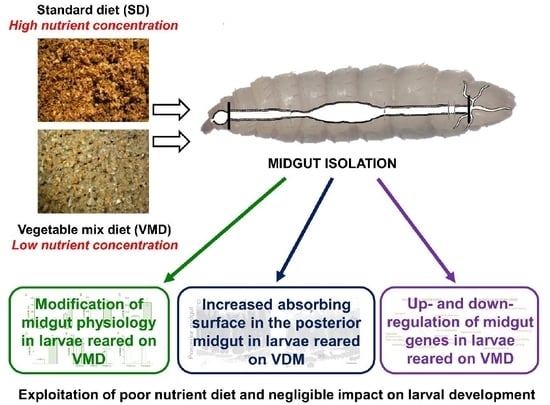
:1. Introduction
2. Results
2.1. Nutrient Content of the Rearing Substrates
2.2. Larval Growth Rate
2.3. pH of Diets and Midgut Lumen
2.4. Enzymatic Assays
2.5. Morphological Analysis of the Larval Midgut
2.6. Histochemical Characterization of the Larval Midgut
2.7. De Novo Transcriptome Analysis
2.8. Differential Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Determination of Nutrient Content of the Diets
4.3. Measurement of Larval Growth Rate
4.4. pH of Diet and Midgut Lumen
4.5. Isolation of Midgut Samples
4.6. Enzymatic Assays
4.7. Statistical Analyses for pH Values, Larval Growth Parameters, and Enzymatic Activities
4.8. Optical Microscopy Analysis of the Midgut Epithelium
4.9. Histochemical Analysis of the Larval Midgut
4.10. RNA Isolation and Illumina Sequencing
4.11. De novo Transcriptome and Functional Annotation
4.12. Differential Gene Expression Analysis
4.13. Availability of Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AHRD | automatic assignment of Human Readable Descriptions pipeline |
| APN | aminopeptidase N |
| BApNA | Nα-Benzoyl-d,l-arginine p-nitroanilide hydrochloride |
| BP | biological processes |
| BSF | black soldier fly |
| P:C ratio | protein:carbohydrate ratio |
| FDR | false discovery rate |
| GO | gene onthology |
| MF | molecular functions |
| NGS | next generation sequencing |
| PAS | periodic acid-Schiff |
| PBS | phosphate buffer saline |
| pNA | p-nitroaniline |
| SAAPPpNA | N-succinyl-Ala–Ala-Pro-Phe p-nitroanilide |
| SD | standard diet |
| TPM | number of transcripts per million |
| VMD | vegetable mix diet |
| U | unit of enzymatic activity |
References
- Clissold, F.J.; Tedder, B.J.; Conigrave, A.D.; Simpson, S.J. The gastrointestinal tract as a nutrient-balancing organ. Proc. R. Soc. B Biol. Sci. 2010, 277, 1751–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.P.; Simpson, S.J.; Clissold, F.J.; Brooks, R.C.; Ballard, J.W.O.; Taylor, P.W.; Soran, N.; Raubenheimer, D. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 2008, 105, 2498–2503. [Google Scholar] [CrossRef] [Green Version]
- Simpson, S.J.; Le Couteur, D.G.; Raubenheimer, D. Putting the Balance Back in Diet. Cell 2015, 161, 18–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solon-Biet, S.M.; McMahon, A.C.; Ballard, J.W.O.; Ruohonen, K.; Wu, L.E.; Cogger, V.C.; Warren, A.; Huang, X.; Pichaud, N.; Melvin, R.G.; et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014, 19, 418–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, S.J.; Raubenheimer, D. Macronutrient balance and lifespan. Aging 2009, 1, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Barragan-Fonseca, K.B.; Dicke, M.; van Loon, J.J.A. Nutritional value of the black soldier fly (Hermetia illucens L.) and its suitability as animal feed—A review. J. Insects Food Feed. 2017, 3, 105–120. [Google Scholar] [CrossRef]
- Raubenheimer, D.; Simpson, S.J. Nutritional ecology and foraging theory. Curr. Opin. Insect Sci. 2018, 27, 38–45. [Google Scholar] [CrossRef]
- Gold, M.; Tomberlin, J.K.; Diener, S.; Zurbrügg, C.; Mathys, A. Decomposition of biowaste macronutrients, microbes, and chemicals in black soldier fly larval treatment: A review. Waste Manag. 2018, 82, 302–318. [Google Scholar] [CrossRef]
- Nguyen, T.T.X.; Tomberlin, J.K.; VanLaerhoven, S. Ability of black soldier fly (Diptera: Stratiomyidae) larvae to recycle food waste. Environ. Entomol. 2015, 44, 406–410. [Google Scholar] [CrossRef]
- Jucker, C.; Erba, D.; Leonardi, M.G.; Lupi, D.; Savoldelli, S. Assessment of vegetable and fruit substrates as potential rearing media for Hermetia illucens (Diptera: Stratiomyidae) larvae. Environ. Entomol. 2017, 46, 1415–1423. [Google Scholar] [CrossRef]
- Spranghers, T.; Ottoboni, M.; Klootwijk, C.; Ovyn, A.; Deboosere, S.; De Meulenaer, B.; Michiels, J.; Eeckhout, M.; De Clercq, P.; De Smet, S. Nutritional composition of black soldier fly (Hermetia illucens) prepupae reared on different organic waste substrates. J. Sci. Food Agric. 2016, 97, 2594–2600. [Google Scholar] [CrossRef]
- Wang, Y.S.; Shelomi, M. Review of black soldier fly (Hermetia illucens) as animal feed and human food. Foods 2017, 6, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bava, L.; Jucker, C.; Gislon, G.; Lupi, D.; Savoldelli, S.; Zucali, M.; Colombini, S. Rearing of Hermetia illucens on different organic by-products: Influence on growth, waste reduction, and environmental impact. Animals 2019, 9, 289. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Rehman, K.U.; Liu, X.; Yang, Q.; Zheng, L.; Li, W.; Cai, M.; Li, Q.; Zhang, J.; Yu, Z. Insect biorefinery: A green approach for conversion of crop residues into biodiesel and protein. Biotechnol. Biofuels 2017, 10, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, M.; Egger, J.; Scheidegger, A.; Zurbrügg, C.; Bruno, D.; Bonelli, M.; Tettamanti, G.; Casartelli, M.; Schmitt, E.; Kerkaert, B.; et al. Estimating black soldier fly larvae biowaste conversion performance by simulation of midgut digestion. Waste Manag. 2020, 112, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Rumpold, B.A.; Klocke, M.; Schlüter, O. Insect biodiversity: Underutilized bioresource for sustainable applications in life sciences. Reg. Environ. Chang. 2016, 17, 1445–1454. [Google Scholar] [CrossRef]
- Vogel, H.; Müller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef]
- Liu, C.; Wang, C.; Yao, H. Comprehensive resource utilization of waste using the black soldier fly (Hermetia illucens (L.)) (Diptera: Stratiomyidae). Animals 2019, 9, 349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbi, S.; Messori, M.; Manfredini, T.; Pini, M.; Montorsi, M. Rational design and characterization of bioplastics from Hermetia illucens prepupae proteins. Biopolymers 2018, 110, e23250. [Google Scholar] [CrossRef]
- Cammack, J.A.; Tomberlin, J.K. The impact of diet protein and carbohydrate on select life-history traits of the black soldier fly Hermetia illucens (L.) (Diptera: Stratiomyidae). Insects 2017, 8, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleissner, D.; Rumpold, B.A. Utilization of organic residues using heterotrophic microalgae and insects. Waste Manag. 2018, 72, 227–239. [Google Scholar] [CrossRef] [PubMed]
- San Martin, D.; Ramos, S.; Zufía, J. Valorization of food waste to produce new raw materials for animal feed. Food Chem. 2016, 198, 68–74. [Google Scholar] [CrossRef]
- Plazzotta, S.; Manzocco, L.; Nicoli, M.C. Fruit and vegetable waste management and the challenge of fresh-cut salad. Trends Food Sci. Technol. 2017, 63, 51–59. [Google Scholar] [CrossRef]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions. Appl. Environ. Microbiol. 2019, 85, 01864–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappellozza, S.; Leonardi, M.G.; Savoldelli, S.; Carminati, D.; Rizzolo, A.; Cortellino, G.; Terova, G.; Moretto, E.; Badaile, A.; Concheri, G.; et al. A first attempt to produce proteins from insects by means of a circular economy. Animals 2019, 9, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Publications Office of the EU; European Commission. Commission Regulation (EU) 2017/893 of 24 May 2017 Amending Annexes I and IV to Regulation (EC) no 999/2001 of the European Parliament and of the Council and Annexes X, XIV and XV to Commission Regulation (EU) no 142/2011 as Regards the Provisions on Processed Animal Protein. Off. J. Eur. Union 2017, L138, 92–116. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32017R0893&from=EN (accessed on 7 July 2020).
- Bonelli, M.; Bruno, D.; Caccia, S.; Sgambetterra, G.; Cappellozza, S.; Jucker, C.; Tettamanti, G.; Casartelli, M. Structural and functional characterization of Hermetia illucens larval midgut. Front. Physiol. 2019, 10, 204. [Google Scholar] [CrossRef]
- Hogsette, J.A. New diets for production of house flies and stable flies (Diptera: Muscidae) in the laboratory. J. Econ. Entomol. 1992, 85, 2291–2294. [Google Scholar] [CrossRef] [Green Version]
- Caccia, S.; Casartelli, M.; Tettamanti, G. The amazing complexity of insect midgut cells: Types, peculiarities, and functions. Cell Tissue Res. 2019, 377, 505–525. [Google Scholar] [CrossRef] [PubMed]
- Callegari, M.; Marasco, R.; Jucker, C.; Mapelli, F.; Fusi, M.; Borin, S.; Daffonchio, D.; Savoldelli, S.; Crotti, E. Developmental stage and diet drive the bacterial community diversity in the food-waste reducing insect Hermetia illucens. In Proceedings of the 4th International Conference on Microbial Diversity “Drivers of Microbial diversity 2017”, Bari, Italy, 24–26 October 2017; pp. 239–242. [Google Scholar]
- Terra, W.R.; Ferreira, C. Insect digestive enzymes: Properties, compartmentalization and function. Comp. Biochem. Physiol. Part B Comp. Biochem. 1994, 109, 1–62. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C.; Jordão, B.P.; Dillon, R.J. Digestive enzymes. In Biology of the Insect Midgut; Springer Science and Business Media LLC: Berlin, Germany, 1996; pp. 153–194. [Google Scholar]
- Nichol, H.; Law, J.H.; Winzerling, J.J. Iron metabolism in insects. Annu. Rev. Entomol. 2002, 47, 535–559. [Google Scholar] [CrossRef]
- Tang, X.; Zhou, B. Iron homeostasis in insects: Insights from Drosophila studies. IUBMB Life 2013, 65, 863–872. [Google Scholar] [CrossRef]
- Mehta, A.; Deshpande, A.; Bettedi, L.; Missirlis, F. Ferritin accumulation under iron scarcity in Drosophila iron cells. Biochimie 2009, 91, 1331–1334. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GitHub. Available online: https://github.com/groupschoof/AHRD (accessed on 7 July 2020).
- NCBI Assembly. Available online: https://www.ncbi.nlm.nih.gov/assembly/ (accessed on 7 July 2020).
- Vicoso, B.; Bachtrog, D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, S.; Fang, G.; Cai, M.; Kou, Z.; Xu, J.; Cao, Y.; Bai, L.; Zhang, Y.; Jiang, Y.; Luo, X.; et al. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res. 2019, 30, 50–60. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Tokuda, G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 2013, 31, 838–850. [Google Scholar] [CrossRef]
- Saadeddin, A. The complexities of hydrolytic enzymes from the termite digestive system. Crit. Rev. Biotechnol. 2012, 34, 115–122. [Google Scholar] [CrossRef]
- Lehane, M.; Blakemore, D.; Williams, S.; Moffatt, M.R. Regulation of digestive enzyme levels in insects. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1995, 110, 285–289. [Google Scholar] [CrossRef]
- Lazarević, J.; Janković-Tomanić, M. Dietary and phylogenetic correlates of digestive trypsin activity in insect pests. Entomol. Exp. Appl. 2015, 157, 123–151. [Google Scholar] [CrossRef]
- Broehan, G.; Kemper, M.; Driemeier, D.; Vogelpohl, I.; Merzendorfer, H. Cloning and expression analysis of midgut chymotrypsin-like proteinases in the tobacco hornworm. J. Insect Physiol. 2008, 54, 1243–1252. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, D.; Zheng, S.; Liu, L.; Tao, S.; Yang, L.; Hu, S.; Feng, Q. A chymotrypsin-like serine protease cDNA involved in food protein digestion in the common cutworm, Spodoptera litura: Cloning, characterization, developmental and induced expression patterns, and localization. J. Insect Physiol. 2010, 56, 788–799. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Zheng, S.; Feng, Q.; Liu, L. A midgut-specific chymotrypsin cDNA (Slctlp1) from Spodoptera litura: Cloning, characterization, localization and expression analysis. Arch. Insect Biochem. Physiol. 2010, 76, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Spit, J.; Zels, S.; Dillen, S.; Holtof, M.; Wynant, N.; Broeck, J.V. Effects of different dietary conditions on the expression of trypsin- and chymotrypsin-like protease genes in the digestive system of the migratory locust, Locusta migratoria. Insect Biochem. Mol. Biol. 2014, 48, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, V. Biosynthesis and control of mosquito gut proteases. IUBMB Life 2003, 55, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Santiago, P.B.; de Araújo, C.N.; Motta, F.N.; Praça, Y.R.; Charneau, S.; Bastos, I.M.D.; Santana, J.M. Proteases of haematophagous arthropod vectors are involved in blood-feeding, yolk formation and immunity—A review. Parasit. Vectors 2017, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Da Lage, J.L. The Amylases of Insects. Int. J. Insect Sci. 2018, 10, 1–14. [Google Scholar] [CrossRef]
- Alfonso, J.; Ortego, F.; Sanchez-Monge, R.; García-Casado, G.; Pujol, M.; Castañera, P.; Salcedo, G. Wheat and barley inhibitors active towards α-Amylase and trypsin-like activities from Spodoptera frugiperda. J. Chem. Ecol. 1997, 23, 1729–1741. [Google Scholar] [CrossRef]
- Kotkar, H.M.; Sarate, P.J.; Tamhane, V.A.; Gupta, V.; Giri, A.P. Responses of midgut amylases of Helicoverpa armigera to feeding on various host plants. J. Insect Physiol. 2009, 55, 663–670. [Google Scholar] [CrossRef]
- Lwalaba, D.; Hoffmann, K.H.; Woodring, J. Control of the release of digestive enzymes in the larvae of the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2009, 73, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Sarate, P.; Tamhane, V.; Kotkar, H.; Ratnakaran, N.; Susan, N.; Gupta, V.; Giri, A.P. Developmental and digestive flexibilities in the midgut of a polyphagous pest, the cotton bollworm, Helicoverpa armigera. J. Insect Sci. 2012, 12, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickey, D.A.; Benkel, B. Regulation of amylase activity in Drosophila melanogaster: Effects of dietary carbohydrate. Biochem. Genet. 1982, 20, 1117–1129. [Google Scholar] [CrossRef]
- Inomata, N.; Kanda, K.; Cariou, M.; Tachida, H.; Yamazaki, T. Evolution of the response patterns to dietary carbohydrates and the developmental differentiation of gene expression of α-amylase in Drosophila. J. Mol. Evol. 1995, 41, 1076–1084. [Google Scholar] [CrossRef]
- Klarenberg, A.J.; Vermeulen, J.W.C.; Jacobs, P.J.M.; Scharloo, W. Genetic and dietary regulation of tissue-specific expression patterns of α-amylase in larvae of Drosophila melanogaster. Comp. Biochem. Physiol. Part B Comp. Biochem. 1988, 89, 143–146. [Google Scholar] [CrossRef]
- Chng, W.A.; Sleiman, M.S.B.; Schüpfer, F.; Lemaitre, B. Transforming Growth Factor β/Activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 2014, 9, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Satake, H.; Takeda, M. Nutrient-induced α-amylase and protease activity is regulated by crustacean cardioactive peptide (CCAP) in the cockroach midgut. Peptides 2006, 27, 2157–2164. [Google Scholar] [CrossRef]
- Weidlich, S.; Müller, S.; Hoffmann, K.H.; Woodring, J. Regulation of amylase, cellulase and chitinase secretion in the digestive tract of the two-spotted field cricket, Gryllus bimaculatus. Arch. Insect Biochem. Physiol. 2013, 83, 69–85. [Google Scholar] [CrossRef]
- Canavoso, L.E.; Jouni, Z.E.; Karnas, K.J.; Pennington, J.E.; Wells, M.A. Fat metabolism in insects. Annu. Rev. Nutr. 2001, 21, 23–46. [Google Scholar] [CrossRef]
- Heier, C.; Kühnlein, R.P. Triacylglycerol metabolism in Drosophila melanogaster. Genetics 2018, 210, 1163–1184. [Google Scholar] [CrossRef] [Green Version]
- Loidl, A.; Crailsheim, K. Free fatty acids digested from pollen and triolein in the honeybee (Apis mellifera carnica Pollmann) midgut. J. Comp. Physiol. B 2001, 171, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Christeller, J.T.; Amara, S.; Carrière, F. Galactolipase, phospholipase and triacylglycerol lipase activities in the midgut of six species of lepidopteran larvae feeding on different lipid diets. J. Insect Physiol. 2011, 57, 1232–1239. [Google Scholar] [CrossRef]
- Weidlich, S.; Hoffmann, K.H.; Woodring, J. Secretion of lipases in the digestive tract of the cricket Gryllus bimaculatus. Arch. Insect Biochem. Physiol. 2015, 90, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, P.W.; Burgess, E.P.J.; Philip, B.A.; McManus, M.T.; Watson, L.; Christeller, J.T. Ultrastructural changes to the midgut of the black field cricket (Teleogryllus commodus) following ingestion of potato protease inhibitor II. J. Insect Physiol. 2002, 48, 327–336. [Google Scholar] [CrossRef]
- Houk, E.I.; Hardy, J.L. Midgut cellular responses to bloodmeal digestion in the mosquito, Culex tarsalis Coquillett (Diptera: Culicidae). Int. J. Insect Morphol. Embryol. 1982, 11, 109–119. [Google Scholar] [CrossRef]
- Rudin, W.; Hecker, H. Functional morphology of the midgut of Aedes aegypti L. (Insecta, Diptera) during blood digestion. Cell Tissue Res. 1979, 200, 193–203. [Google Scholar] [CrossRef]
- Franzetti, E.; Romanelli, D.; Caccia, S.; Cappellozza, S.; Congiu, T.; Rajagopalan, M.; Grimaldi, A.; de Eguileor, M.; Casartelli, M.; Tettamanti, G. The midgut of the silkmoth Bombyx mori is able to recycle molecules derived from degeneration of the larval midgut epithelium. Cell Tissue Res. 2015, 361, 509–528. [Google Scholar] [CrossRef]
- Yamada, T.; Habara, O.; Yoshii, Y.; Matsushita, R.; Kubo, H.; Nojima, Y.; Nishimura, T. The role of glycogen in development and adult fitness in Drosophila. Development 2019, 146, dev176149. [Google Scholar] [CrossRef] [Green Version]
- Bruno, D.; Bonelli, M.; Cadamuro, A.G.; Reguzzoni, M.; Grimaldi, A.; Casartelli, M.; Tettamanti, G. The digestive system of the adult Hermetia illucens (Diptera: Stratiomyidae): Morphological features and functional properties. Cell Tissue Res. 2019, 378, 221–238. [Google Scholar] [CrossRef]
- Turunen, S.; Crailsheim, K. Lipid and sugar absorption. In Biology of the Insect Midgut; Springer Science and Business Media LLC: Berlin, Germany, 1996; pp. 293–320. [Google Scholar]
- Buchon, N.; Osman, D.; David, F.P.; Fang, H.Y.; Boquete, J.P.; Deplancke, B.; Lemaitre, B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013, 3, 1725–1738. [Google Scholar] [CrossRef] [Green Version]
- Marianes, A.; Spradling, A.C. Physiological and stem cell compartmentalization within the Drosophila midgut. eLife 2013, 2, 00886. [Google Scholar] [CrossRef]
- Ruaud, A.F.; Lam, G.; Thummel, C.S. The Drosophila NR4A nuclear receptor DHR38 regulates carbohydrate metabolism and glycogen storage. Mol. Endocrinol. 2011, 25, 83–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, J.H. Insects, oxygen, and iron. Biochem. Biophys. Res. Commun. 2002, 292, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Missirlis, F.; Kosmidis, S.; Brody, T.; Mavrakis, M.; Holmberg, S.; Odenwald, W.F.; Skoulakis, E.M.C.; Rouault, T.A. Homeostatic mechanisms for iron storage revealed by genetic manipulations and live imaging of Drosophila ferritin. Genetics 2007, 177, 89–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, D.V. Molecular and genetic analyses of Drosophila prat, which encodes the first enzyme of de novo purine biosynthesis. Genetics 1994, 136, 547–557. [Google Scholar] [PubMed]
- Chávez, V.M.; Marqués, G.; Delbecque, J.P.; Kobayashi, K.; Hollingsworth, M.; Burr, J.E.; Natzle, J.; O’Connor, M.B. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 2000, 127, 4115–4126. [Google Scholar]
- Warren, J.T.; Petryk, A.; Marqués, G.; Jarcho, M.; Parvy, J.P.; Dauphin-Villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef] [Green Version]
- Navarro, J.A.; Ohmann, E.; Sanchez, D.; Botella, J.A.; Liebisch, G.; Moltó, M.D.; Ganfornina, M.D.; Schmitz, G.; Schneuwly, S. Altered lipid metabolism in a Drosophila model of Friedreich’s ataxia. Hum. Mol. Genet. 2010, 19, 2828–2840. [Google Scholar] [CrossRef] [Green Version]
- Pham, D.Q.D.; Winzerling, J.J. Insect ferritins: Typical or atypical? Biochim. Biophys. Acta Gen. Subj. 2010, 1800, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Mandilaras, K.; Pathmanathan, T.; Missirlis, F. Iron absorption in Drosophila melanogaster. Nutrients 2013, 5, 1622–1647. [Google Scholar] [CrossRef] [Green Version]
- Filshie, B.K.; Poulson, D.F.; Waterhouse, D.F. Ultrastructure of the copper-accumulating region of the Drosophila larval midgut. Tissue Cell 1971, 3, 77–102. [Google Scholar] [CrossRef]
- Poulson, D.F.; Bowen, V.T. Organization and function of the inorganic constituents of nuclei. Exp. Cell Res. 1952, 2, 161–180. [Google Scholar]
- Pimentel, A.C.; Montali, A.; Bruno, D.; Tettamanti, G. Metabolic adjustment of the larval fat body in Hermetia illucens to dietary conditions. J. Asia-Pacific Entomol. 2017, 20, 1307–1313. [Google Scholar] [CrossRef]
- Horwitz, W. AOAC International Official Methods of Analysis of AOAC International, 17th ed.; The Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2000. [Google Scholar]
- Latimer, G.W. AOAC International Official Methods of Analysis of AOAC International, 20th ed.; The Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2016. [Google Scholar]
- Van Soest, P.V.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Charney, J.; Tomarelli, R.M. A colorimetric method for the determination of the proteolytic activity of duodenal juice. J. Biol. Chem. 1947, 171, 501–505. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 7 July 2020).
- FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 7 July 2020).
- Bushnell, B. BBMap. 2015. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 7 July 2020).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology Consortium; Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.E.; Lewis, S.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2018, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ENA European Nucleotide Archive. Available online: https://www.ebi.ac.uk/ena (accessed on 7 July 2020).










| Standard Diet | Vegetable Mix Diet | |||
|---|---|---|---|---|
| Component | As Fed | Dry Matter | As Fed | Dry Matter |
| Crude protein | 6.4 | 14.1 | 1.2 | 10.3 |
| Crude lipid | 1.2 | 2.7 | 0.1 | 0.7 |
| Crude fiber a | 4.9 | 10.8 | 0.5 | 4.4 |
| Nitrogen-free extract b | 30.2 | 67.3 | 9.6 | 80.0 |
| Ash | 2.3 | 5.1 | 0.6 | 4.6 |
| Hemicellulose c | 9.8 | 21.3 | 0.4 | 3.6 |
| Cellulose c | 4.4 | 9.7 | 0.6 | 4.6 |
| Lignin c | 1.7 | 3.7 | 0.2 | 1.3 |
| Starch | 8.5 | 18.8 | 1.4 | 11.6 |
| Glucose and fructose | 1.5 | 3.3 | 1.5 | 12.8 |
| Moisture | 55 | - | 88 | - |
| Standard Diet | Vegetable Mix Diet | |||
|---|---|---|---|---|
| Component | As Fed | Dry Matter | As Fed | Dry Matter |
| Iron | 119.1 | 261.8 | 3.1 | 26.9 |
| Copper | 4.2 | 9.3 | 0.8 | 6.8 |
| Nickel | 0.8 | 1.7 | 0.1 | 0.7 |
| Zinc | 22.4 | 49.3 | 2.1 | 18.1 |
| Moisture | 55 | - | 88 | - |
| SD | VMD | |
|---|---|---|
| Anterior midgut | 5.8 ± 0.1 (6) | 6.0 ± 0.1 (6) |
| Middle midgut | 2.4 ± 0.2 (6) | 1.8 ± 0.2 (7) |
| Posterior midgut | 8.3 ± 0.4 (6) | 8.8 ± 0.1 (7) |
| Sequencing and Assembly Parameters | Value |
|---|---|
| Total number of transcripts a | 27,102 |
| Minimum length (nt) | 201 |
| Maximum length (nt) | 26,717 |
| Average length (nt) | 1170 |
| N50 of transcripts (nt) b | 2137 |
| Reads remapped (%) | 86.81 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonelli, M.; Bruno, D.; Brilli, M.; Gianfranceschi, N.; Tian, L.; Tettamanti, G.; Caccia, S.; Casartelli, M. Black Soldier Fly Larvae Adapt to Different Food Substrates through Morphological and Functional Responses of the Midgut. Int. J. Mol. Sci. 2020, 21, 4955. https://doi.org/10.3390/ijms21144955
Bonelli M, Bruno D, Brilli M, Gianfranceschi N, Tian L, Tettamanti G, Caccia S, Casartelli M. Black Soldier Fly Larvae Adapt to Different Food Substrates through Morphological and Functional Responses of the Midgut. International Journal of Molecular Sciences. 2020; 21(14):4955. https://doi.org/10.3390/ijms21144955
Chicago/Turabian StyleBonelli, Marco, Daniele Bruno, Matteo Brilli, Novella Gianfranceschi, Ling Tian, Gianluca Tettamanti, Silvia Caccia, and Morena Casartelli. 2020. "Black Soldier Fly Larvae Adapt to Different Food Substrates through Morphological and Functional Responses of the Midgut" International Journal of Molecular Sciences 21, no. 14: 4955. https://doi.org/10.3390/ijms21144955
APA StyleBonelli, M., Bruno, D., Brilli, M., Gianfranceschi, N., Tian, L., Tettamanti, G., Caccia, S., & Casartelli, M. (2020). Black Soldier Fly Larvae Adapt to Different Food Substrates through Morphological and Functional Responses of the Midgut. International Journal of Molecular Sciences, 21(14), 4955. https://doi.org/10.3390/ijms21144955