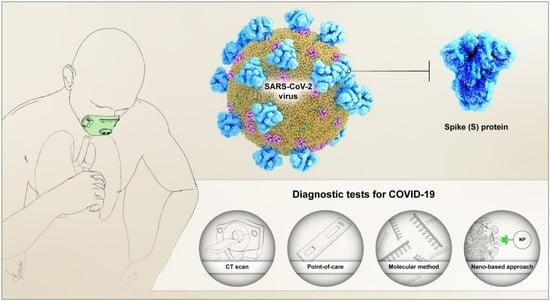
Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic
Abstract
:1. Introduction
2. Characteristics of SARS-CoV-2 towards the Possible Mechanism of Diagnosis and Treatment
3. Current Diagnostic Techniques for the Detection of SARS-CoV-2
3.1. Isothermal Nucleic Acid Amplification
3.2. Microarray-Based Technologies
3.3. CRISPR Technology
3.4. Point-of-Care Detection of SARS-CoV-2
4. Nanotechnology to Detect SARS-CoV-2
4.1. Based on Gold NPs
4.2. Based on Ag NPs
4.3. Based on Magnetic NPs
4.4. Based on Metal-Organic Framework
5. Conclusion and Future Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Word Health Organization. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). Available online: https://www.who.int/publications/i/item/report-of-the-who-china-joint-mission-on-coronavirus-disease-2019-(covid-19) (accessed on 15 June 2020).
- Baud, D.; Qi, X.; Nielsen-Saines, K.; Musso, D.; Pomar, L.; Favre, G. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020, 20, 773. [Google Scholar] [CrossRef] [Green Version]
- Farassati, F. Signal-smart oncolytic viruses in treatment of human cancers. U.S. Patent 16/198, 578, 23 May 2019. [Google Scholar]
- Haller, O.; Weber, F. Pathogenic viruses: Smart manipulators of the interferon system. In Interferon: The 50th Anniversary; Springer: Berlin, Germany, 2007; pp. 315–334. [Google Scholar]
- García, M.; Cooper, A.; Shi, W.; Bornmann, W.; Carrion, R.; Kalman, D.; Nabel, G.J. Productive replication of Ebola virus is regulated by the c-Abl1 tyrosine kinase. Sci. Transl. Med. 2012, 4, 123ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Word Health Organization. Coronavirus disease 2019 (COVID-19): Situation report, 59. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 1 July 2020).
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020, 200642. [Google Scholar] [CrossRef] [Green Version]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR test results in patients recovered from COVID-19. Jama 2020, 323, 1502–1503. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.-Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. Jama 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020. [Google Scholar] [CrossRef]
- Remuzzi, A.; Remuzzi, G. COVID-19 and Italy: What next? Lancet 2020, 395, 1225–1228. [Google Scholar] [CrossRef]
- Arabi, Y.M.; Murthy, S.; Webb, S. COVID-19: A novel coronavirus and a novel challenge for critical care. Intensive Care Med. 2020, 46, 1–4. [Google Scholar]
- Casadevall, A.; Pirofski, L.-a. The convalescent sera option for containing COVID-19. J. Clin. Investig. 2020, 130, 1545–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourouiba, L. Turbulent Gas Clouds and Respiratory Pathogen Emissions: Potential Implications for Reducing Transmission of COVID-19. Jama 2020, 323, 1837–1838. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Du, R.; Wang, R.; Cao, T.; Guan, L.; Yang, C.; Zhu, Q.; Hu, M.; Li, X.; Li, Y. Comparison of Hospitalized Patients with Acute Respiratory Distress Syndrome Caused by COVID-19 and H1N1. Chest 2020, 158, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Lipsitch, M.; Swerdlow, D.L.; Finelli, L. Defining the epidemiology of Covid-19—Studies needed. New Engl. J. Med. 2020, 382, 1194–1196. [Google Scholar] [CrossRef]
- Tebyetekerwa, M.; Xu, Z.; Yang, S.; Ramakrishna, S. Electrospun Nanofibers-Based Face Masks. Adv. Fiber Mater. 2020. [Google Scholar] [CrossRef]
- Chagas, A.M.; Molloy, J.C.; Prieto-Godino, L.L.; Baden, T. Leveraging Open Hardware to alleviate the burden of COVID-19 on global health systems. Plos Biol. 2020, 18, 3–6. [Google Scholar]
- Jernigan, D.B. Update: Public health response to the coronavirus disease 2019 outbreak—United States, February 24, 2020. Mmwr. Morb. Mortal. Wkly. Rep. 2020, 69, 216–219. [Google Scholar] [CrossRef]
- Yu, L.; Wu, S.; Hao, X.; Li, X.; Liu, X.; Ye, S.; Han, H.; Dong, X.; Li, X.; Li, J. Rapid colorimetric detection of COVID-19 coronavirus using a reverse tran-scriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic plat-form: ILACO. medRxiv 2020. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Yang, Z. Can a Paper-Based Device Trace COVID-19 Sources with Wastewater-Based Epidemiology? Environ. Sci. Technol. 2020, 54, 3733–3735. [Google Scholar] [CrossRef]
- Nundy, S.; Patel, K.K. Self-Service Diagnosis of COVID-19—Ready for Prime Time? In Proceedings of the JAMA Health Forum, Plymouth, PA, USA, 2 March 2020; American Medial Association: Chicago, IL, USA, 2020; p. e200333. [Google Scholar]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, H.; Pöhlmann, S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004, 12, 466–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Siddique, R.; Shereen, M.A.; Ali, A.; Liu, J.; Bai, Q.; Bashir, N.; Xue, M. The emergence of a novel coronavirus (SARS-CoV-2), their biology and therapeutic options. J. Clin. Microbiol. 2020, 58, 187–200. [Google Scholar] [CrossRef] [Green Version]
- Huynh, K. Reduced hospital admissions for ACS—more collateral damage from COVID-19. Nat. Rev. Cardiol. 2020, 17, 453. [Google Scholar] [CrossRef]
- Eastman, R.T.; Roth, J.S.; Brimacombe, K.R.; Simeonov, A.; Shen, M.; Patnaik, S.; Hall, M.D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. Acs Cent. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Li, G.; Xu, F.; Zhang, Z. Metal–organic frameworks: Opportunities and challenges for surface-enhanced Raman scattering–a review. J. Mater. Chem. C 2020, 8, 2952–2963. [Google Scholar] [CrossRef]
- Aguilera-Sigalat, J.; Bradshaw, D. Synthesis and applications of metal-organic framework–quantum dot (QD@ MOF) composites. Coord. Chem. Rev. 2016, 307, 267–291. [Google Scholar] [CrossRef] [Green Version]
- Zhan, G.; Zeng, H.C. Integrated nanocatalysts with mesoporous silica/silicate and microporous MOF materials. Coord. Chem. Rev. 2016, 320, 181–192. [Google Scholar] [CrossRef]
- Falcaro, P.; Ricco, R.; Yazdi, A.; Imaz, I.; Furukawa, S.; Maspoch, D.; Ameloot, R.; Evans, J.D.; Doonan, C.J. Application of metal and metal oxide nanoparticles@ MOFs. Coord. Chem. Rev. 2016, 307, 237–254. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sexton, N.R.; Smith, E.C.; Blanc, H.; Vignuzzi, M.; Peersen, O.B.; Denison, M.R. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016, 90, 7415–7428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020. [Google Scholar] [CrossRef] [Green Version]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C. Diagnosing COVID-19: The disease and tools for detection. Acs Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guobao, F.; Lu, L.; Wanzhao, C.; Wang, F. Electron Beam Irradiation on Novel Coronavirus (COVID-19): Via a Monte-Carlo Simulation. Chin. Phys. B 2020. [Google Scholar] [CrossRef]
- Monajjemi, M.; Mollaamin, F.; Shojaei, S. An overview on Coronaviruses family from past to Covid-19: Introduce some inhibitors as antiviruses from Gillan’s plants. Biointerface Res. Appl. Chem. 2020, 10, 5575–5585. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Zhao, Y.-B.; Wang, Q.; Li, J.-Y.; Zhou, Z.-J.; Liao, C.-H.; Ge, X.-Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020. [Google Scholar] [CrossRef]
- Gurwitz, D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med Virol. 2020, 92, 903–908. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, 58. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiol. 2020, 200432. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.; Zerunian, M.; Polici, M.; Pucciarelli, F.; Polidori, T.; Rucci, C.; Guido, G.; Bracci, B.; De Dominicis, C.; Laghi, A. Chest CT Features of COVID-19 in Rome, Italy. Radiol. 2020, 201237. [Google Scholar] [CrossRef]
- Li, Y.; Xia, L. Coronavirus disease 2019 (COVID-19): Role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020, 214, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Cao, Q.; Qin, L.; Wang, X.; Cheng, Z.; Pan, A.; Dai, J.; Sun, Q.; Zhao, F.; Qu, J. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): A multi-center study in Wenzhou city, Zhejiang, China. J. Infect. 2020, 80, 388–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Bacon, J.W.; Jasti, R. Gram-scale synthesis and crystal structures of [8]-and [10] CPP, and the solid-state structure of C 60@[10] CPP. Chem. Sci. 2012, 3, 3018–3021. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, M.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, P.R.; Sethy, K.; Mohapatra, S.; Panda, D. Loop mediated isothermal amplification: An innovative gene amplification technique for animal diseases. Vet. World 2016, 9, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chander, Y.; Koelbl, J.; Puckett, J.; Moser, M.J.; Klingele, A.J.; Liles, M.R.; Carrias, A.; Mead, D.A.; Schoenfeld, T.W. A novel thermostable polymerase for RNA and DNA loop-mediated isothermal amplification (LAMP). Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Wang, H.; Cao, Z.; Jin, H.; Chi, H.; Zhao, J.; Yu, B.; Yan, F.; Hu, X.; Wu, F.; et al. A Rapid and Specific Assay for the Detection of MERS-CoV. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kang, M.; Park, E.; Chung, D.R.; Kim, J.; Hwang, E.S. A Simple and Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of SARS-CoV. Biochip J. 2019, 13, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, Y.; Ge, Y.; Zhao, K.; Zou, B.; Liu, B.; Qi, X.; Bian, Q.; Shi, Z.; Zhu, F.; Zhou, M.; et al. Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Cascade Invasive Reaction and Nanoparticle Hybridization for Subtyping of Influenza A Virus. Sci. Rep. 2017, 7, 44924. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Odiwuor, N.; Xiong, J.; Sun, L.; Nyaruaba, R.O.; Wei, H.; Tanner, N.A. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. medRxiv 2020. [Google Scholar] [CrossRef]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) Assays Targeting SARS-CoV-2. J. Mol. Diagn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; Nguyen, T.; Chromikova, V.; Strohmeier, S.; Stadlbauer, D.; Javier, A.; Jiang, K.; Asthagiri-Arunkumar, G.; Polanco, J.; Bermudez-Gonzalez, M.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv 2020. [Google Scholar] [CrossRef]
- Wang, K.; Yee, C.-C.; Au-Yeung, H.Y. Facile syntheses of [3]-,[4]-and [6] catenanes templated by orthogonal supramolecular interactions. Chem. Sci. 2016, 7, 2787–2792. [Google Scholar] [CrossRef] [Green Version]
- Fernando, I.R.; Frasconi, M.; Wu, Y.; Liu, W.G.; Wasielewski, M.R.; Goddard, W.A., 3rd; Stoddart, J.F. Sliding-Ring Catenanes. J. Am. Chem. Soc. 2016, 138, 10214–10225. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Soon, W.W.; Hariharan, M.; Snyder, M.P. High-throughput sequencing for biology and medicine. Mol. Syst. Biol. 2013, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardinge, P.; Murray, J.A.H. Reduced False Positives and Improved Reporting of Loop-Mediated Isothermal Amplification using Quenched Fluorescent Primers. Sci. Rep. 2019, 9, 7400. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Poon, L.L.; Leung, C.S.; Tashiro, M.; Chan, K.H.; Wong, B.W.; Yuen, K.Y.; Guan, Y.; Peiris, J.S. Rapid detection of the severe acute respiratory syndrome (SARS) coronavirus by a loop-mediated isothermal amplification assay. Clin. Chem. 2004, 50, 1050–1052. [Google Scholar] [CrossRef] [Green Version]
- Notomi, T.; Taguchi, F.; Kanda, H.; Minekawa, H.; Itamura, S.; Odagiri, T.; Tashiro, M. RT-LAMP method provides a simple, rapid and specific detection system for SARS-CoV RNA. In Proceedings of the International Conference on SARS-one year after the (first) outbreak, Lübeck, Germany, 11 May 2004; German Medical Science: Düsseldorf, Germany, 2004. [Google Scholar]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Lee, S.H.; Baek, Y.H.; Kim, Y.-H.; Choi, Y.-K.; Song, M.-S.; Ahn, J.-Y. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front. Microbiol. 2017, 7, 2166. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Cong, F.; Zeng, F.; Lian, Y.; Liu, X.; Luo, M.; Guo, P.; Ma, J. Development of a real time reverse transcription loop-mediated isothermal amplification method (RT-LAMP) for detection of a novel swine acute diarrhea syndrome coronavirus (SADS-CoV). J. Virol. Methods 2018, 260, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Thai, H.T.C.; Le, M.Q.; Vuong, C.D.; Parida, M.; Minekawa, H.; Notomi, T.; Hasebe, F.; Morita, K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004, 42, 1956–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, M.; Zhou, Y.; Ye, J.; AL-maskri, A.A.A.; Kang, Y.; Zeng, S.; Cai, S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020, 10, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Semba, S.; El-Kafrawy, S.A.; Hassan, A.M.; Tolah, A.M.; Takayama, I.; Kageyama, T.; Notomi, T.; Kamitani, W.; Matsuyama, S. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J. Virol. Methods 2018, 258, 41–48. [Google Scholar] [CrossRef]
- Jiang, Y.S.; Bhadra, S.; Li, B.; Wu, Y.R.; Milligan, J.N.; Ellington, A.D. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015, 87, 3314–3320. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, S.; Jiang, Y.S.; Kumar, M.R.; Johnson, R.F.; Hensley, L.E.; Ellington, A.D. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV). PLoS ONE 2015, 10, 123126. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Wang, B.; Potter, S.J.; Lin, Y.; Cunningham, A.L.; Dwyer, D.E.; Su, Y.; Ma, X.; Hou, Y.; Saksena, N.K. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J. Clin. Microbiol. 2005, 43, 2339–2344. [Google Scholar] [CrossRef] [Green Version]
- Ciftci, S.; Neumann, F.; Abdurahman, S.; Appelberg, K.S.; Mirazimi, A.; Nilsson, M.; Madaboosi, N. Digital Rolling Circle Amplification–Based Detection of Ebola and Other Tropical Viruses. J. Mol. Diagn. 2020, 22, 272–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.; Nakajima, R.; Jain, A.; De Assis, R.R.; Jasinskas, A.; Obiero, J.M.; Adenaiye, O.; Tai, S.; Hong, F.; Milton, D.K. Analysis of Serologic Cross-Reactivity Between Common Human Coronaviruses and SARS-CoV-2 Using Coronavirus Antigen Microarray. BioRxiv 2020. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7239054/pdf/nihpp-2020.03.24.006544.pdf (accessed on 25 March 2020). [CrossRef] [Green Version]
- Wang, D.; Urisman, A.; Liu, Y.-T.; Springer, M.; Ksiazek, T.G.; Erdman, D.D.; Mardis, E.R.; Hickenbotham, M.; Magrini, V.; Eldred, J. Viral discovery and sequence recovery using DNA microarrays. Plos Biol. 2003, 1, e2. [Google Scholar] [CrossRef] [Green Version]
- de Souza Luna, L.K.; Heiser, V.; Regamey, N.; Panning, M.; Drexler, J.F.; Mulangu, S.; Poon, L.; Baumgarte, S.; Haijema, B.J.; Kaiser, L. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J. Clin. Microbiol. 2007, 45, 1049–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Geng, P.; Wang, Q.; Cao, B.; Liu, B. Development of a Single Nucleotide Polymorphism DNA Microarray for the Detection and Genotyping of the SARS Coronavirus. J. Microbiol. Biotechnol. 2014, 24, 1445–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juang, J.-L.; Chen, T.-C.; Jiang, S.S.; Hsiung, C.A.; Chen, W.-C.; Chen, G.-W.; Lin, S.-M.; Lin, J.-H.; Chiu, S.-C.; Lai, Y.-K. Coupling multiplex RT-PCR to a gene chip assay for sensitive and semiquantitative detection of severe acute respiratory syndrome-coronavirus. Lab. Investig. 2004, 84, 1085–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chertow, D.S. Next-generation diagnostics with CRISPR. Science 2018, 360, 381–382. [Google Scholar] [CrossRef]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 2019, 76, 826–837.e811. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, W.; Fasching, C.L.; Singh, J.; Chiu, C.Y.; Chen, J.S. A protocol for rapid detection of the 2019 novel coronavirus SARS-CoV-2 using CRISPR diagnostics: SARS-CoV-2 DETECTR. Available online: https://mammoth.bio/wp-content/uploads/2020/04/200423-A-protocol-for-rapid-detection-of-SARS-CoV-2-using-CRISPR-diagnostics_3.pdf (accessed on 23 April 2020).
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-based DETECTR Lateral Flow Assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Liu, C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV virus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.E.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.; Debs, R. Development of CRISPR as a prophylactic strategy to combat novel coronavirus and influenza. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Lucia, C.; Federico, P.-B.; Alejandra, G.C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y. Development and Evaluation of A CRISPR-based Diagnostic For 2019-novel Coronavirus. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. New Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, L.; Li, F.; Han, Y.L.; Lin, M.; Lu, T.J.; Xu, F. Oligonucleotide-linked gold nanoparticle aggregates for enhanced sensitivity in lateral flow assays. Lab A Chip 2013, 13, 4352–4357. [Google Scholar] [CrossRef] [PubMed]
- Dalirirad, S.; Steckl, A.J. Lateral flow assay using aptamer-based sensing for on-site detection of dopamine in urine. Anal. Biochem. 2020, 596, 113637. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Seo, H.S.; Kwon, J.-H.; Kim, H.-T.; Kwon, K.C.; Sim, S.J.; Cha, Y.J.; Lee, J. Multiplex diagnosis of viral infectious diseases (AIDS, hepatitis C, and hepatitis A) based on point of care lateral flow assay using engineered proteinticles. Biosens. Bioelectron. 2015, 69, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Ha, K.; Guk, K.; Lee, D.K.; Eom, G.; Song, S.; Kang, T.; Park, H.; Jung, J.; Lim, E.-K. Rapid and simple detection of Tamiflu-resistant influenza virus: Development of oseltamivir derivative-based lateral flow biosensor for point-of-care (POC) diagnostics. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, J.; Hu, J.; Long, Q.; Deng, H.; Fan, K.; Liao, P.; Liu, B.; Wu, G.; Chen, Y. A Peptide-based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Corona Virus Disease 2019 (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Sugikawa, K.; Nagata, S.; Furukawa, Y.; Kokado, K.; Sada, K. Stable and functional gold nanorod composites with a metal–organic framework crystalline shell. Chem. Mater. 2013, 25, 2565–2570. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.A.I.; Henry, E.; Ringe, R.P.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Schierhorn, M.; Lee, S.J.; Boettcher, S.W.; Stucky, G.D.; Moskovits, M. Metal–Silica Hybrid Nanostructures for Surface-Enhanced Raman Spectroscopy. Adv. Mater. 2006, 18, 2829–2832. [Google Scholar] [CrossRef]
- Sawai, Y.; Takimoto, B.; Nabika, H.; Ajito, K.; Murakoshi, K. Observation of a small number of molecules at a metal nanogap arrayed on a solid surface using surface-enhanced Raman scattering. J. Am. Chem. Soc. 2007, 129, 1658–1662. [Google Scholar] [CrossRef] [PubMed]
- Zhan, W.-w.; Kuang, Q.; Zhou, J.-z.; Kong, X.-j.; Xie, Z.-x.; Zheng, L.-s. Semiconductor@ metal–organic framework core–shell heterostructures: A case of ZnO@ ZIF-8 nanorods with selective photoelectrochemical response. J. Am. Chem. Soc. 2013, 135, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C. Nano Research for COVID-19. Acs Nano 2020, 14, 3719–3720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; He, Y.; Yang, X.; Yuan, R.; Chai, Y. Porous SiO2@ Ni@ C and Au nanocages as surface-enhanced Raman spectroscopy platform with use of DNA structure switching for sensitive detection of uracil DNA glycolase. Sens. Actuators B: Chem. 2020, 304, 127273. [Google Scholar] [CrossRef]
- Shi, P.; Liu, Z.; Dong, K.; Ju, E.; Ren, J.; Du, Y.; Li, Z.; Qu, X. A Smart “Sense-Act-Treat” System: Combining a Ratiometric pH Sensor with a Near Infrared Therapeutic Gold Nanocage. Adv. Mater. 2014, 26, 6635–6641. [Google Scholar] [CrossRef]
- Wang, L.-J.; Chang, Y.-C.; Sun, R.; Li, L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens. Bioelectron. 2017, 87, 686–692. [Google Scholar] [CrossRef]
- Soler, M.; Huertas, C.S.; Lechuga, L.M. Label-free plasmonic biosensors for point-of-care diagnostics: A review. Expert Rev. Mol. Diagn. 2019, 19, 71–81. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Laing, S.; Gracie, K.; Faulds, K. Multiplex in vitro detection using SERS. Chem. Soc. Rev. 2016, 45, 1901–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emonds-Alt, G.; Mignolet, B.; Malherbe, C.; Monbaliu, J.-C.M.; Remacle, F.; Eppe, G. Understanding chemical interaction between phosphonate-derivative molecules and a silver surface cluster in SERS: A combined experimental and computational approach. Phys. Chem. Chem. Phys. 2019, 21, 22180–22187. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yan, H.; Xue, X.; Jiang, D.; Cai, Y.; Liang, D.; Jung, Y.M.; Han, X.X.; Zhao, B. Surface-Enhanced Raman Scattering (SERS) Active Gold Nanoparticles Decorated on a Porous Polymer Filter. Appl. Spectrosc. 2017, 71, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Al-Shalalfeh, M.M.; Saleh, T.A.; Al-Saadi, A.A. Silver colloid and film substrates in surface-enhanced Raman scattering for 2-thiouracil detection. Rsc Adv. 2016, 6, 75282–75292. [Google Scholar] [CrossRef]
- Boginskaya, I.; Sedova, M.; Baburin, A.; Afanas’ev, K.; Zverev, A.; Echeistov, V.; Ryzhkov, V.; Rodionov, I.; Tonanaiskii, B.; Ryzhikov, I.; et al. SERS-Active Substrates Nanoengineering Based on e-Beam Evaporated Self-Assembled Silver Films. Appl. Sci. 2019, 9, 3988. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.-T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.-W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “lab-on-a-chip” to detect viruses–Current challenges and future perspectives. Biosens. Bioelectron. 2020, 112291. [Google Scholar] [CrossRef]
- Zhao, Z.; Cui, H.; Song, W.; Ru, X.; Zhou, W.; Yu, X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mosier-Boss, A.P. Review of SERS Substrates for Chemical Sensing. Nanomaterials 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; Wu, D.Y.; et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef]
- Lin, X.-D.; Uzayisenga, V.; Li, J.-F.; Fang, P.-P.; Wu, D.-Y.; Ren, B.; Tian, Z.-Q. Synthesis of ultrathin and compact Au@MnO2 nanoparticles for shell-isolated nanoparticle-enhanced Raman spectroscopy (SHINERS). J. Raman Spectrosc. 2012, 43, 40–45. [Google Scholar] [CrossRef]
- Liu, X.; Wang, J.; Wu, Y.; Fan, T.; Xu, Y.; Tang, L.; Ying, Y. Compact Shielding of Graphene Monolayer Leads to Extraordinary SERS-Active Substrate with Large-Area Uniformity and Long-Term Stability. Sci. Rep. 2015, 5, 17167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirsadeghi, S.; Dinarvand, R.; Ghahremani, M.H.; Hormozi-Nezhad, M.R.; Mahmoudi, Z.; Hajipour, M.J.; Atyabi, F.; Ghavami, M.; Mahmoudi, M. Protein corona composition of gold nanoparticles/nanorods affects amyloid beta fibrillation process. Nanoscale 2015, 7, 5004–5013. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, F.; Hormozi-Nezhad, M.R.; Mahmoudi, M. A colorimetric sensor array for detection and discrimination of biothiols based on aggregation of gold nanoparticles. Anal. Chim. Acta 2015, 882, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Nejad, M.A.F.; Bigdeli, A.; Hormozi-Nezhad, M.R. Wide color-varying visualization of sulfide with a dual emissive ratiometric fluorescence assay using carbon dots and gold nanoclusters. Microchem. J. 2020, 104960. [Google Scholar] [CrossRef]
- Moslehipour, A.; Bigdeli, A.; Ghasemi, F.; Hormozi-Nezhad, M.R. Design of a ratiometric fluorescence nanoprobe to detect plasma levels of levodopa. Microchem. J. 2019, 148, 591–596. [Google Scholar] [CrossRef]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; de la Guardia, M. Nanomaterial-based biosensors for detection of pathogenic virus. Trac Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Draz, M.S.; Shafiee, H. Applications of gold nanoparticles in virus detection. Theranostics 2018, 8, 1985. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Kim, G.H.; Kim, S.M.; Hong, K.; Kim, Y.; Park, C.; Sohn, H.; Min, J. Label-free localized surface plasmon resonance biosensor composed of multi-functional DNA 3 way junction on hollow Au spike-like nanoparticles (HAuSN) for avian influenza virus detection. Colloids Surf. B Biointerfaces 2019, 182, 110341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Pang, D.W.; Zhang, Z.L.; Zheng, H.Z.; Cao, J.P.; Shen, J.T. Visual gene diagnosis of HBV and HCV based on nanoparticle probe amplification and silver staining enhancement. J. Med Virol. 2003, 70, 205–211. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Shen, J.-T.; Liu, H.-H. Analytical performance of and real sample analysis with an HBV gene visual detection chip. J. Virol. Methods 2004, 121, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Dong, X.; Zhang, K.; Han, X.; Fang, X.; Zhang, Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst 2013, 138, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, O.; Morita, M.; Kato, T.; Ito, M.; Suzuki, T.; Park, E.Y. Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays. Biosens. Bioelectron. 2017, 94, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cushing, S.K.; Liang, H.; Suri, S.; Ma, D.; Wu, N. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal. Chem. 2013, 85, 2072–2078. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Li, H.; Xu, D. Enzyme-free fluorescence microarray for determination of hepatitis B virus DNA based on silver nanoparticle aggregates-assisted signal amplification. Anal. Chim. Acta 2019, 1077, 297–304. [Google Scholar] [CrossRef]
- Hung, L.-Y.; Chang, J.-C.; Tsai, Y.-C.; Huang, C.-C.; Chang, C.-P.; Yeh, C.-S.; Lee, G.-B. Magnetic nanoparticle-based immunoassay for rapid detection of influenza infections by using an integrated microfluidic system. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 819–829. [Google Scholar] [CrossRef] [PubMed]
- de la Escosura-Muñiz, A.; Maltez-da Costa, M.; Sánchez-Espinel, C.; Díaz-Freitas, B.; Fernández-Suarez, J.; González-Fernández, Á.; Merkoçi, A. Gold nanoparticle-based electrochemical magnetoimmunosensor for rapid detection of anti-hepatitis B virus antibodies in human serum. Biosens. Bioelectron. 2010, 26, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Liu, J.; Saha, R.; Su, D.; Krishna, V.D.; Cheeran, M.C.; Wang, J.-P. Detection of Influenza A Virus Nucleoprotein Through the Self-Assembly of Nanoparticles in Magnetic Particle Spectroscopy-Based Bioassays: A Method for Rapid, Sensitive, and Wash-free Magnetic Immunoassays. arXiv 2019, arXiv:1907.06000. [Google Scholar]
- Zhang, Q.; Wang, C.-F.; Lv, Y.-K. Luminescent switch sensors for the detection of biomolecules based on metal–organic frameworks. Analyst 2018, 143, 4221–4229. [Google Scholar] [CrossRef]
- Ploetz, E.; Engelke, H.; Lächelt, U.; Wuttke, S. The Chemistry of Reticular Framework Nanoparticles: MOF, ZIF, and COF Materials. Adv. Funct. Mater. 2020, 1909062. [Google Scholar] [CrossRef]
- Xie, B.-P.; Qiu, G.-H.; Hu, P.-P.; Liang, Z.; Liang, Y.-M.; Sun, B.; Bai, L.-P.; Jiang, Z.-H.; Chen, J.-X. Simultaneous detection of Dengue and Zika virus RNA sequences with a three-dimensional Cu-based zwitterionic metal–organic framework, comparison of single and synchronous fluorescence analysis. Sens. Actuators B Chem. 2018, 254, 1133–1140. [Google Scholar] [CrossRef]
- Wei, X.; Zheng, L.; Luo, F.; Lin, Z.; Guo, L.; Qiu, B.; Chen, G. Fluorescence biosensor for the H 5 N 1 antibody based on a metal–organic framework platform. J. Mater. Chem. B 2013, 1, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-P.; Chen, S.-R.; Liu, S.-W.; Tang, X.-Y.; Qin, L.; Qiu, G.-H.; Chen, J.-X.; Chen, W.-H. Platforms formed from a three-dimensional Cu-based zwitterionic metal–organic framework and probe ss-DNA: Selective fluorescent biosensors for human immunodeficiency virus 1 ds-DNA and Sudan virus RNA sequences. Anal. Chem. 2015, 87, 12206–12214. [Google Scholar] [CrossRef]
- Jia, Z.; Ma, Y.; Yang, L.; Guo, C.; Zhou, N.; Wang, M.; He, L.; Zhang, Z. NiCo2O4 spinel embedded with carbon nanotubes derived from bimetallic NiCo metal-organic framework for the ultrasensitive detection of human immune deficiency virus-1 gene. Biosens. Bioelectron. 2019, 133, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.F.; Fang, R.M.; Huang, C.Z.; Li, Y.F. Dual amplifying fluorescence anisotropy for detection of respiratory syncytial virus DNA fragments with size-control synthesized metal–organic framework MIL-101. Rsc Adv. 2015, 5, 46301–46306. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Baran, T.; Baran, N.Y.; Sajjadi, M.; Tahsili, M.R.; Shokouhimehr, M. Pd nanocatalyst stabilized on amine-modified zeolite: Antibacterial and catalytic activities for environmental pollution remediation in aqueous medium. Sep. Purif. Technol. 2020, 239, 116542. [Google Scholar] [CrossRef]
- Le, Q.V.; Yang, G.; Wu, Y.; Jang, H.W.; Shokouhimehr, M.; Oh, Y.K. Nanomaterials for modulating innate immune cells in cancer immunotherapy. Asian J. Pharm. Sci. 2019, 14, 16–29. [Google Scholar] [CrossRef]
- Beitollahi, H.; Tajik, S.; Dourandish, Z.; Zhang, K.; Le, Q.V.; Jang, H.W.; Kim, S.Y.; Shokouhimehr, M. Recent Advances in the Aptamer-Based Electrochemical Biosensors for Detecting Aflatoxin B1 and Its Pertinent Metabolite Aflatoxin M1. Sensors 2020, 20, 3256. [Google Scholar] [CrossRef]








| Methods | Sensitivity of Detection | Analyses of Time | Advantages | Limitation | Ref |
|---|---|---|---|---|---|
| Culture | 30–50% | 1–3 days | Appropriated for slower-growing pathogens | Risk of contamination, time-consuming | [46] |
| Next-generation sequencing (NGS) | - | Around 1–2 weeks | Ability to fully recognize the genome, even mutations. Detection based on genes | Time-consuming, short reads, need for technical expertise | [67,68] |
| Immunoassays Methods (e.g., ELISA) | 20–80% | About 2 h with the kit | High sensitivity, ability to detect IgG and IgM antibodies in serum. Detection based on antibodies | Expensive to prepare antibody, limit the amount of antigen in samples, antibody instability | [69] |
| RT-PCR | 95% | 2 h | Highly sensitive method | Requires expensive instruments, long reaction times | [22,55] |
| LAMP | >95% | About 30 min | Rapid, simple, high specificity and specificity | Difficult in primer design, false-positive results | [62,70] |
| Computed tomography (CT) | 97% | Rapid | Rapid analysis | Non-specific | [8] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabiee, N.; Bagherzadeh, M.; Ghasemi, A.; Zare, H.; Ahmadi, S.; Fatahi, Y.; Dinarvand, R.; Rabiee, M.; Ramakrishna, S.; Shokouhimehr, M.; et al. Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 5126. https://doi.org/10.3390/ijms21145126
Rabiee N, Bagherzadeh M, Ghasemi A, Zare H, Ahmadi S, Fatahi Y, Dinarvand R, Rabiee M, Ramakrishna S, Shokouhimehr M, et al. Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic. International Journal of Molecular Sciences. 2020; 21(14):5126. https://doi.org/10.3390/ijms21145126
Chicago/Turabian StyleRabiee, Navid, Mojtaba Bagherzadeh, Amir Ghasemi, Hossein Zare, Sepideh Ahmadi, Yousef Fatahi, Rassoul Dinarvand, Mohammad Rabiee, Seeram Ramakrishna, Mohammadreza Shokouhimehr, and et al. 2020. "Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic" International Journal of Molecular Sciences 21, no. 14: 5126. https://doi.org/10.3390/ijms21145126
APA StyleRabiee, N., Bagherzadeh, M., Ghasemi, A., Zare, H., Ahmadi, S., Fatahi, Y., Dinarvand, R., Rabiee, M., Ramakrishna, S., Shokouhimehr, M., & Varma, R. S. (2020). Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic. International Journal of Molecular Sciences, 21(14), 5126. https://doi.org/10.3390/ijms21145126